Team:UC Davis/Notebook/Week 7
From 2011.igem.org
| Line 140: | Line 140: | ||
color:#EE3333; | color:#EE3333; | ||
font-size: 20px; | font-size: 20px; | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
} | } | ||
Revision as of 07:42, 28 September 2011
Start a Family
Got a favorite BioBrick? Check our our process for expanding basic parts into part families.Criteria
View our judging criteria for iGEM 2011 here.
 Construction is getting very close to being done after this week! We are having to do a lot of PCR screens due to the amount of background on our vector controls, but it looks like we have most of our repressors in front of B0034 and most of our promoters in front of GFP and pBad. We're just starting on putting these two intermediate parts together! We have also began redeveloping our mutagenic PCR protocol and plan to test it first on GFP.
Construction is getting very close to being done after this week! We are having to do a lot of PCR screens due to the amount of background on our vector controls, but it looks like we have most of our repressors in front of B0034 and most of our promoters in front of GFP and pBad. We're just starting on putting these two intermediate parts together! We have also began redeveloping our mutagenic PCR protocol and plan to test it first on GFP.
--Monday 7/25/11--
We continued with construction today and, once again, extracted the cut Promoters+GFP and repressors from a gel. We ligated the promoters+GFP in from of I13453 and the repressors before B0034. Hopefully this is the last time we will have to ligate and transform these parts.
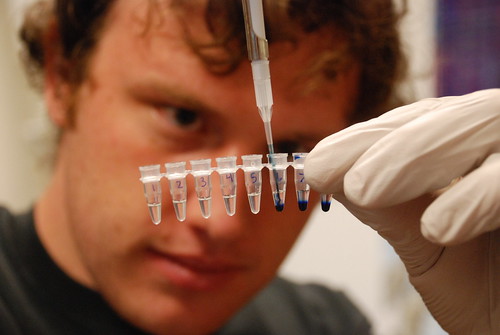
We decided to develop a new mutagenic PCR protocol. Drawing from this paper, we tweaked the MnCl2, MgCl2, dNTP, and template DNA concentrations, while scaling the reaction volume to 50 uL.
In total, eight different reaction conditions across a range of MgCl2 and MnCl2 values were prepared for testing with R0010 (lacI-repressible promoter).

The insert controls for yesterday's transformations looked good, but there were still a good amount of colonies on the vector controls. We decided to do a PCR screen of these parts to make sure that they were the right length.
Today was also "Tip Day"! We filled enough tip boxes to last us a quite a while.
PCR concentrations from mutagenic PCR were low. We ran them again with much higher template concentrations (10X and 100X).
We decided it was time to revamp our error-prone PCR protocol. Using this paper as a guide, we created 16 different protocols, with the intention of picking the one that produces the most viable mutants.
Today we continued construction. We're almost done with our testing construct and have are making headway with producing our mutants.
We cut: R0051+E0240+I13453 and R0040+E0240+I13453 with Eco and Spe in order to put B0034+C0051 and B0034+C0040 downstream by cutting with Eco and Xba. We also cut E0240+I13453 and B0034+C0012 so that we could put them together for use in the LacI testing construct.
As usual we're tinkering with our wiki and constantly updating it with new content.
Nanodrop concentrations for PCR products reported yesterday were done on the wrong tubes (!). Concentrations at 0.1 uL were fine.
We extracted the digestions from yesterday but got poor concentrations so we did a fast digest so that we could come in over the weekend and hopefully have successful ligations by Monday.
Began testing mutagenic PCR on longer fragments (GFP); we hope to judge mutation rate by visual inspection using GFP as template.
We ligated R0040+E0240+I13453 in front of both B0034+C0040 and B0034, and ligated R0051+E0240+I13453 in front of both B0034+C0051 and B0034. We then transformed these products, along with their controls, onto carb plates.
 "
"




