Team:WashU/Notebook/Transformation
From 2011.igem.org
(→Sept. 24) |
J.herrmann (Talk | contribs) (→Transformation Group) |
||
| (6 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
== Transformation Group == | == Transformation Group == | ||
| - | + | [[File:Yeast Transformation.jpg|300px|thumb|right|]] | |
| - | The team | + | The goal of the transformation team was to successfully integrate the four synthesized genes into the yeast genome. |
| + | Once the genes are ligated into a construct with their respective selection markers, we will use homology regions from the yeast genome to integrate the linear piece of DNA into the yeast genome. After inserting one gene per yeast cell, we will mate MATa and MATalpha strains yeast to make two diploid strains, each containing two of our genes of interest. We will then sporulate the two strains and select for a yeast strain that has integrated all four genes into its genome. | ||
| - | + | Additionally, this team transformed plasmids into e. coli in order to amplify plasmids for use by the other teams. This procedure consisted of transforming the plasmid into the bacterial genome, culturing the transformed bacteria, and isolating the DNA plasmid by means of a mini-prep kit. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:Bacteria Plate.jpg|400px|thumb|center|]] | |
Latest revision as of 03:47, 28 September 2011
Transformation Group
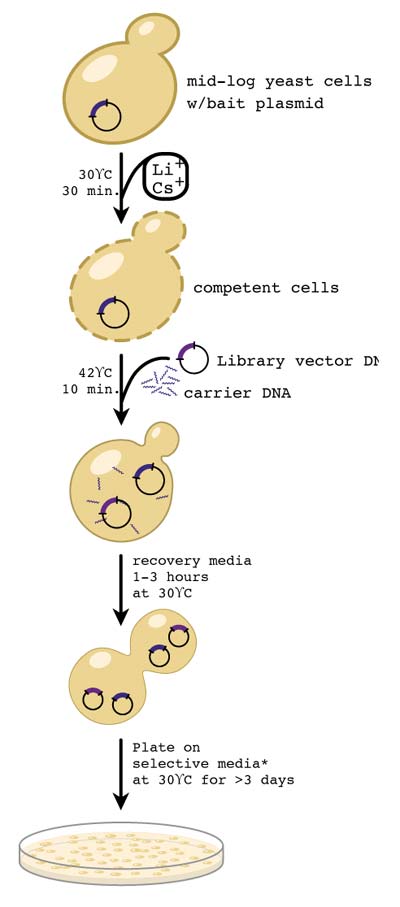
The goal of the transformation team was to successfully integrate the four synthesized genes into the yeast genome.
Once the genes are ligated into a construct with their respective selection markers, we will use homology regions from the yeast genome to integrate the linear piece of DNA into the yeast genome. After inserting one gene per yeast cell, we will mate MATa and MATalpha strains yeast to make two diploid strains, each containing two of our genes of interest. We will then sporulate the two strains and select for a yeast strain that has integrated all four genes into its genome.
Additionally, this team transformed plasmids into e. coli in order to amplify plasmids for use by the other teams. This procedure consisted of transforming the plasmid into the bacterial genome, culturing the transformed bacteria, and isolating the DNA plasmid by means of a mini-prep kit.
 "
"