Team:BU Wellesley Software/SBOL
From 2011.igem.org
| (17 intermediate revisions not shown) | |||
| Line 45: | Line 45: | ||
</style> | </style> | ||
<link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/Team.css"> | <link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/Team.css"> | ||
| + | <script type="text/javascript"> | ||
| + | </script> | ||
</head> | </head> | ||
| Line 97: | Line 99: | ||
<div id="overview"> | <div id="overview"> | ||
<h1>SBOL Overview</h1> | <h1>SBOL Overview</h1> | ||
| - | <a href="http://www.sbolstandard.org/">SBOL</a> | + | <p> |
| - | <br><center><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/SBOL/SBOL1.png" | + | The Synthetic Biology Open Language <a href="http://www.sbolstandard.org/">(SBOL)</a> is an emerging standard which attempts to describe the information used in synthetic biological designs. SBOL has two components: SBOL semantic and SBOL visual. Semantic captures the organization and types of data comprising a design. Visual provides iconography for biological concepts such as promoters, ribosome binding sites, and terminators. SBOL is a community driven standard and a provides a governance board. SBOL attempts to be extensible so that it can grow to meet the needs of the community. It is important that software tools begin to incorporate SBOL in order to begin the process of unifying the synthetic biology design community and better understand the types of information that need to be passed from design to design.</p> |
| + | <br><center><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/SBOL/SBOL1.png" height="300" width="675"> <p><i>SBOL Overview. Here we see both the visual iconography as well as the semantic standard by which a design is decomposed into components and features.</i></center> | ||
<br> | <br> | ||
</div> | </div> | ||
<div id="results"> | <div id="results"> | ||
<h1>Plan</h1> | <h1>Plan</h1> | ||
| - | <br><center><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/SBOL/SBOL2.png" > <p><i>Tools communicate via SBOL</i></center> | + | <p> |
| + | In our project we incorporated SBOL by using the Clotho design environment. Clotho supports SBOL via its "Hermes" app. Hermes is compliant with the libSBOL distribution (Java implementation of the SBOL Core Data Model) <a href="https://github.com/synbiodex">(link)</a>. Hermes allows for both the import and export of data in SBOL. For example, a design in Clotho comprising Clotho Features and Clotho Parts can be exported to SBOL. The process is "lossy" however as SBOL does not currently support all of Clotho's core object's information. Likewise SBOL data can be imported into Clotho to create new Clotho Feature and Part objects. This process is "underspecified" as again not all information for the various Clotho objects' instantiation is provided by SBOL. The missing data defaults to pre-specified values currently. | ||
| + | |||
| + | This process is powerful because it will allow Clotho to share designs with tools such as Tinkercell, GenoCAD, BioFAB data access client, and iBioSim.</p> | ||
| + | <br><center><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/SBOL/SBOL2.png" height="450" width="750" > <p><i>Tools communicate via SBOL. Together tools can now be chained into workflows to provide specific design support and customized algorithms.</i></center> | ||
<br> | <br> | ||
</div> | </div> | ||
<div id="futurework"> | <div id="futurework"> | ||
<h1>Future Work</h1> | <h1>Future Work</h1> | ||
| - | <br><center><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/SBOL/SBOL3.png" > <p><i>SBOL is a continuing collaboration between a variety of research groups.</i></center> | + | <p> |
| + | The key currently is to find a way to make sure SBOL becomes more expressive so that the process of information transfer between Clotho and SBOL is more compatible where appropriate. We also need to "round trip" designs from tool to tool and from wet lab to wet lab. This will require that we take designs experimentally created and validated at Boston University, capture these in Clotho, export the design via Hermes, and then pass this design to another lab using either Clotho or another SBOL compatible tool. That lab will then need to recreate the Boston University results with minimal additional information or intervention from BU. In much the same way electronic design can go from specification to synthesis to assembly with simple Verilog design files, SBOL should enable the same concepts for biology.</p> | ||
| + | |||
| + | <br><center><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/SBOL/SBOL3.png" height="372" width="687" > <p><i>SBOL is a continuing collaboration between a variety of research groups. These collaborations will grow and become more rich in the future.</i></center> | ||
<br> | <br> | ||
</div> | </div> | ||
Latest revision as of 00:19, 28 September 2011
SBOL Integration
SBOL Overview
The Synthetic Biology Open Language (SBOL) is an emerging standard which attempts to describe the information used in synthetic biological designs. SBOL has two components: SBOL semantic and SBOL visual. Semantic captures the organization and types of data comprising a design. Visual provides iconography for biological concepts such as promoters, ribosome binding sites, and terminators. SBOL is a community driven standard and a provides a governance board. SBOL attempts to be extensible so that it can grow to meet the needs of the community. It is important that software tools begin to incorporate SBOL in order to begin the process of unifying the synthetic biology design community and better understand the types of information that need to be passed from design to design.
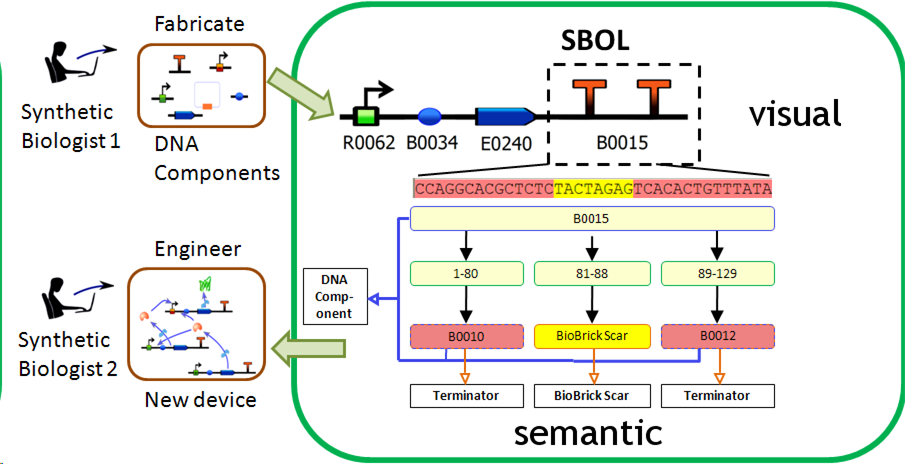
SBOL Overview. Here we see both the visual iconography as well as the semantic standard by which a design is decomposed into components and features.
Plan
In our project we incorporated SBOL by using the Clotho design environment. Clotho supports SBOL via its "Hermes" app. Hermes is compliant with the libSBOL distribution (Java implementation of the SBOL Core Data Model) (link). Hermes allows for both the import and export of data in SBOL. For example, a design in Clotho comprising Clotho Features and Clotho Parts can be exported to SBOL. The process is "lossy" however as SBOL does not currently support all of Clotho's core object's information. Likewise SBOL data can be imported into Clotho to create new Clotho Feature and Part objects. This process is "underspecified" as again not all information for the various Clotho objects' instantiation is provided by SBOL. The missing data defaults to pre-specified values currently. This process is powerful because it will allow Clotho to share designs with tools such as Tinkercell, GenoCAD, BioFAB data access client, and iBioSim.
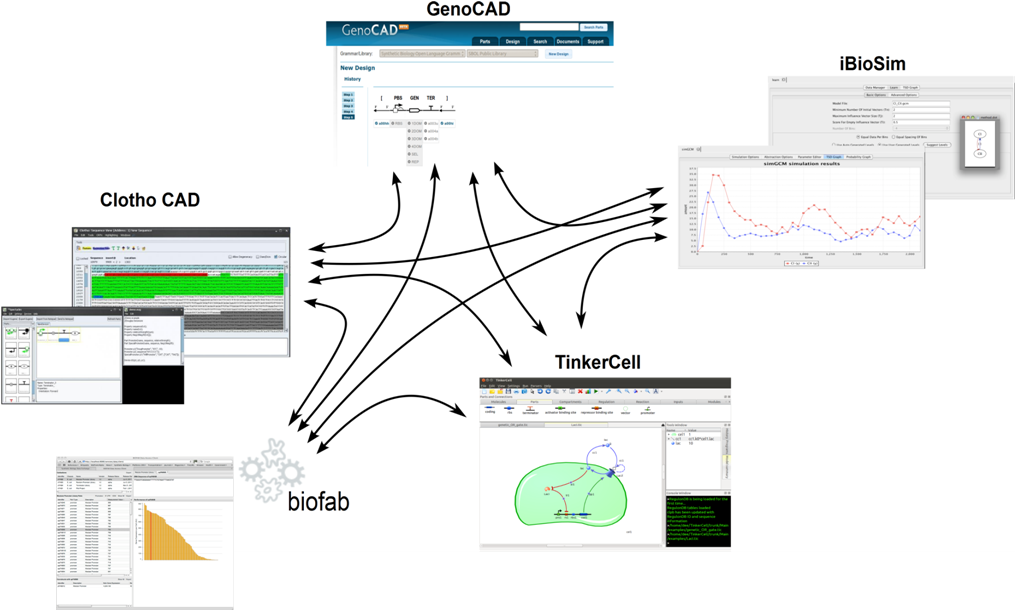
Tools communicate via SBOL. Together tools can now be chained into workflows to provide specific design support and customized algorithms.
Future Work
The key currently is to find a way to make sure SBOL becomes more expressive so that the process of information transfer between Clotho and SBOL is more compatible where appropriate. We also need to "round trip" designs from tool to tool and from wet lab to wet lab. This will require that we take designs experimentally created and validated at Boston University, capture these in Clotho, export the design via Hermes, and then pass this design to another lab using either Clotho or another SBOL compatible tool. That lab will then need to recreate the Boston University results with minimal additional information or intervention from BU. In much the same way electronic design can go from specification to synthesis to assembly with simple Verilog design files, SBOL should enable the same concepts for biology.
SBOL is a continuing collaboration between a variety of research groups. These collaborations will grow and become more rich in the future.
 "
"
