RecA Project
From 2011.igem.org
(Difference between revisions)
| Line 44: | Line 44: | ||
<center><img src="https://2011.igem.org/File:RecA_strategy.jpg" height="300px"/></center> | <center><img src="https://2011.igem.org/File:RecA_strategy.jpg" height="300px"/></center> | ||
<p> | <p> | ||
| + | |||
| + | [[File:RecA strategy.JPG|500px|The above figure shows RecA1 as the blue band, a strand of DNA. The first round of mutations results in a plasmid that can be used in assembly. The second round of mutations results in three different mutated sites in three different plasmids. The third round of mutagenesis adds a mutated site to each of the three plasmids. A final round of mutations will result in a plasmid containing all five mutated sites, the activated RecA gene.]] | ||
Revision as of 05:42, 27 September 2011

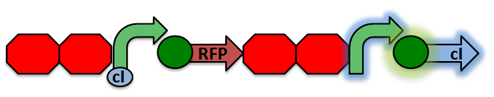
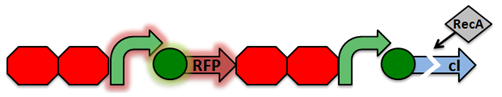
RecA Project Sensor Project Reporter Project
RecA Co-Protease
The normal laboratory strain of E. coli, DH10B, contains an inactivated form of RecA known as RecA1. Therefore, a series of mutations must occur in order to restore its proteatic function and reduce recombination.
Activation of RecA
- Point Mutation at b.p 720: A ⇒ G
Removal of Recombinase Activity
- Arginine 243 ⇒ Glutamine
- Lysine 286 ⇒ Asparagine
In order to use the standard bio-bricking techniques, we also had to remove these naturally occurring enzymatic restriction sites
- PST1 Site [CTGCAG]
- CAG ⇒ CTG (Codon usage bias decreases: .69 to .31)
- EcoR1 Site [GAATTC]
- TTC ⇒ TTT (Codon usage bias increases: .49 to .51)
Activated RecA then cleaves the cI repressor, causing transcription of RFP. However, if RecA exhibits recombinant activity, then the homologous double terminator regions will recombine. This causes the deletion of the repressible promoter and RFP.
 "
"