Team:UCL London/Research/Supercoiliology/Results
From 2011.igem.org
| (11 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
<h1>Results</h1> | <h1>Results</h1> | ||
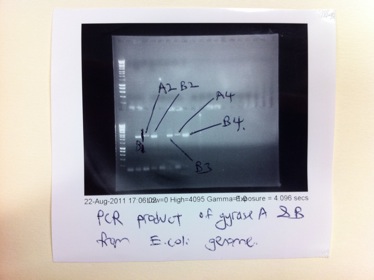
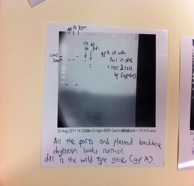
| - | <h2>Extraction and amplification of | + | <h2>Extraction and amplification of GyrA (2627bp) and GyrB (2414bp) genes from the E.coli genome:</h2> |
[[File:Pic1_gyr.jpg]] | [[File:Pic1_gyr.jpg]] | ||
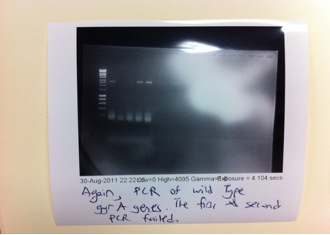
| + | From the PCR reactions above, we have successfully obtained quite a good concentration of GyrB but the PCR of GyrA did not yield a better concentration of the product. Since PCR purification will cause a substantial loss of PCR product (we only obtained about 6-10 ng/μl) we had decided to do a second round of PCR to amplify GyrA. | ||
| - | + | [[File:Pic2_gyr.jpg]] [[File:Gyr3.jpg]] [[File:Gyr4.jpg]] | |
| - | + | ||
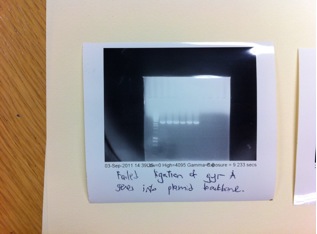
| - | From the transformation which we made to amplify the RFP expression cassette such as BBa_J04450 (pSB1C3,pSB1A3,pSB1K3 plasmid backbones) , we obtained a good concentration of the plasmid DNA from our | + | From the transformation which we made to amplify the RFP expression cassette such as BBa_J04450 (pSB1C3,pSB1A3,pSB1K3 plasmid backbones), we obtained a good concentration of the plasmid DNA from our mini-prep about 130-188 ng/μl. As shown in the gel, the plasmids contain the correct insert which is the RFP cassette of about 1069bp in length (available in BBa_J14460). |
| + | [[File:Gyr5.jpg]] [[File:Gyr6.jpg]] | ||
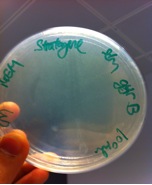
We have also transform our competent TOP10 E.coli cells with the potential inducible promoter such as the ara promoter as well as the IPTG inducible promoter, the RBS and the terminator (BBa_B0015). | We have also transform our competent TOP10 E.coli cells with the potential inducible promoter such as the ara promoter as well as the IPTG inducible promoter, the RBS and the terminator (BBa_B0015). | ||
| Line 24: | Line 25: | ||
<h2>Ligations of PCR products into pSB1C3 , transformation and digestion to inspect the recombinant plasmid inserts:</h2> | <h2>Ligations of PCR products into pSB1C3 , transformation and digestion to inspect the recombinant plasmid inserts:</h2> | ||
| - | Initially we had done many attempts of ligations | + | Initially we had done many attempts of ligations and unfortunately most of them failed. After protocols trouble shooting and gene sequencing, we found out that most of the failure were mostly due to the problems we had encountered with the linearised plasmid backbones (not with the parts as we initially suspected) which were included together with the kit. We then move on and check another possible alternative such as using the BBa_J04450 plasmids to ligate our parts. Luckily, the 3A assembly of using the plasmid succeed and we move on to ligate our PCR products into the pSB1C3 backbone. |
| + | |||
| + | [[File:Gyr7.jpg]] | ||
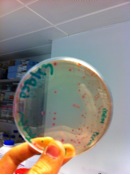
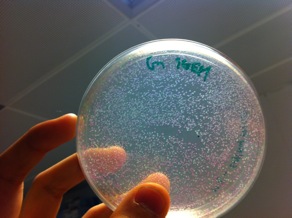
The above photo is one of the successful ligation where the red colonies indicating that the pSB1C3 ligated back to the RFP expression cassette while the white colonies are most likely to have the correct inserts. | The above photo is one of the successful ligation where the red colonies indicating that the pSB1C3 ligated back to the RFP expression cassette while the white colonies are most likely to have the correct inserts. | ||
| + | [[File:Gyr8.jpg]] | ||
| - | The ligations of GyrA into the plasmid backbone from the BBa_J14450 have failed and fortunately for GyrB we successfully ligated the PCR products into the pSB1C3 backbone. We also have a fairly good concentration from the | + | The ligations of GyrA into the plasmid backbone from the BBa_J14450 have failed and fortunately for GyrB we successfully ligated the PCR products into the pSB1C3 backbone. We also have a fairly good concentration from the mini-prep of the 3 overnight cultures (of which the 2nd ,3rd and 5th overnight cultures had failed). The concentration of DNA we have is about 150-165 ng/μl. |
| + | |||
| + | [[File:Gyr9.jpg]] | ||
The following failed attempts for the ligations of GyrA genes into the plasmid backbones. | The following failed attempts for the ligations of GyrA genes into the plasmid backbones. | ||
| - | In our other attempts , we have used TOPO Blunt end cloning kit to try to ligate our GyrA PCR product into the a TOPO plasmid backbone. The plasmid backbone appeared to be ligated back to itself when we digested the | + | In our other attempts, we have used TOPO Blunt end cloning kit to try to ligate our GyrA PCR product into the a TOPO plasmid backbone. The plasmid backbone appeared to be ligated back to itself when we digested the mini-prep plasmids. |
<h2>Site directed mutagenesis of Gyr genes:</h2> | <h2>Site directed mutagenesis of Gyr genes:</h2> | ||
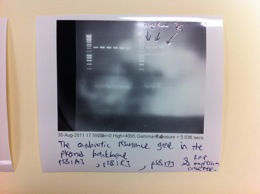
| - | Moving on , we had decided to do site directed mutagenesis of GyrB in the pSB1C3 alone . We have use the | + | Moving on, we had decided to do site directed mutagenesis of GyrB in the pSB1C3 alone. We have use the stratagene SDM kit to carry out the intended mutagenesis to remove the EcoR1 restriction site at the 6th bp position in the GyrB gene. The colonies which we obtained from the stratagene kit were different compared to the ones we got from the site directed mutagenesis of GyrB with the SDM we did with Phusion enzymes. We repeated our SDM several times but unfortunately we failed to obtain the desire products when we check the plasmids through digestion and gel electrophoresis. |
| - | + | ||
| - | Due to the amount of time we have left until the wiki freeze deadline as well as advises from our supervisors, we have decided to put the gyrase project to rest while focusing on our GBS project as well as the | + | [[File:Gyr10.jpg]] [[File:Gyr11.jpg]] |
| - | Please do refer to the GBS project for our | + | |
| + | Due to the amount of time we have left until the wiki freeze deadline as well as advises from our supervisors, we have decided to put the gyrase project to rest while focusing on our GBS project as well as the characterisation of our GBS Biobricks (fermentation and fluorescence measurement). | ||
| + | Please do refer to the GBS project for our characterisation and experimental results. | ||
</div> | </div> | ||
{{:Team:UCL_London/Template/Footer}} | {{:Team:UCL_London/Template/Footer}} | ||
Latest revision as of 04:55, 22 September 2011
Contents |
Results
Extraction and amplification of GyrA (2627bp) and GyrB (2414bp) genes from the E.coli genome:
From the PCR reactions above, we have successfully obtained quite a good concentration of GyrB but the PCR of GyrA did not yield a better concentration of the product. Since PCR purification will cause a substantial loss of PCR product (we only obtained about 6-10 ng/μl) we had decided to do a second round of PCR to amplify GyrA.
From the transformation which we made to amplify the RFP expression cassette such as BBa_J04450 (pSB1C3,pSB1A3,pSB1K3 plasmid backbones), we obtained a good concentration of the plasmid DNA from our mini-prep about 130-188 ng/μl. As shown in the gel, the plasmids contain the correct insert which is the RFP cassette of about 1069bp in length (available in BBa_J14460).
We have also transform our competent TOP10 E.coli cells with the potential inducible promoter such as the ara promoter as well as the IPTG inducible promoter, the RBS and the terminator (BBa_B0015).
Ligations of PCR products into pSB1C3 , transformation and digestion to inspect the recombinant plasmid inserts:
Initially we had done many attempts of ligations and unfortunately most of them failed. After protocols trouble shooting and gene sequencing, we found out that most of the failure were mostly due to the problems we had encountered with the linearised plasmid backbones (not with the parts as we initially suspected) which were included together with the kit. We then move on and check another possible alternative such as using the BBa_J04450 plasmids to ligate our parts. Luckily, the 3A assembly of using the plasmid succeed and we move on to ligate our PCR products into the pSB1C3 backbone.
The above photo is one of the successful ligation where the red colonies indicating that the pSB1C3 ligated back to the RFP expression cassette while the white colonies are most likely to have the correct inserts.
The ligations of GyrA into the plasmid backbone from the BBa_J14450 have failed and fortunately for GyrB we successfully ligated the PCR products into the pSB1C3 backbone. We also have a fairly good concentration from the mini-prep of the 3 overnight cultures (of which the 2nd ,3rd and 5th overnight cultures had failed). The concentration of DNA we have is about 150-165 ng/μl.
The following failed attempts for the ligations of GyrA genes into the plasmid backbones. In our other attempts, we have used TOPO Blunt end cloning kit to try to ligate our GyrA PCR product into the a TOPO plasmid backbone. The plasmid backbone appeared to be ligated back to itself when we digested the mini-prep plasmids.
Site directed mutagenesis of Gyr genes:
Moving on, we had decided to do site directed mutagenesis of GyrB in the pSB1C3 alone. We have use the stratagene SDM kit to carry out the intended mutagenesis to remove the EcoR1 restriction site at the 6th bp position in the GyrB gene. The colonies which we obtained from the stratagene kit were different compared to the ones we got from the site directed mutagenesis of GyrB with the SDM we did with Phusion enzymes. We repeated our SDM several times but unfortunately we failed to obtain the desire products when we check the plasmids through digestion and gel electrophoresis.
Due to the amount of time we have left until the wiki freeze deadline as well as advises from our supervisors, we have decided to put the gyrase project to rest while focusing on our GBS project as well as the characterisation of our GBS Biobricks (fermentation and fluorescence measurement). Please do refer to the GBS project for our characterisation and experimental results.
 "
"