Team:Wageningen UR/Project/ModelingProj1
From 2011.igem.org
(→Modeling synchronized oscillations) |
(→Modeling synchronized oscillations) |
||
| (One intermediate revision not shown) | |||
| Line 28: | Line 28: | ||
{{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ | {{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ | ||
=== Mathematical model === | === Mathematical model === | ||
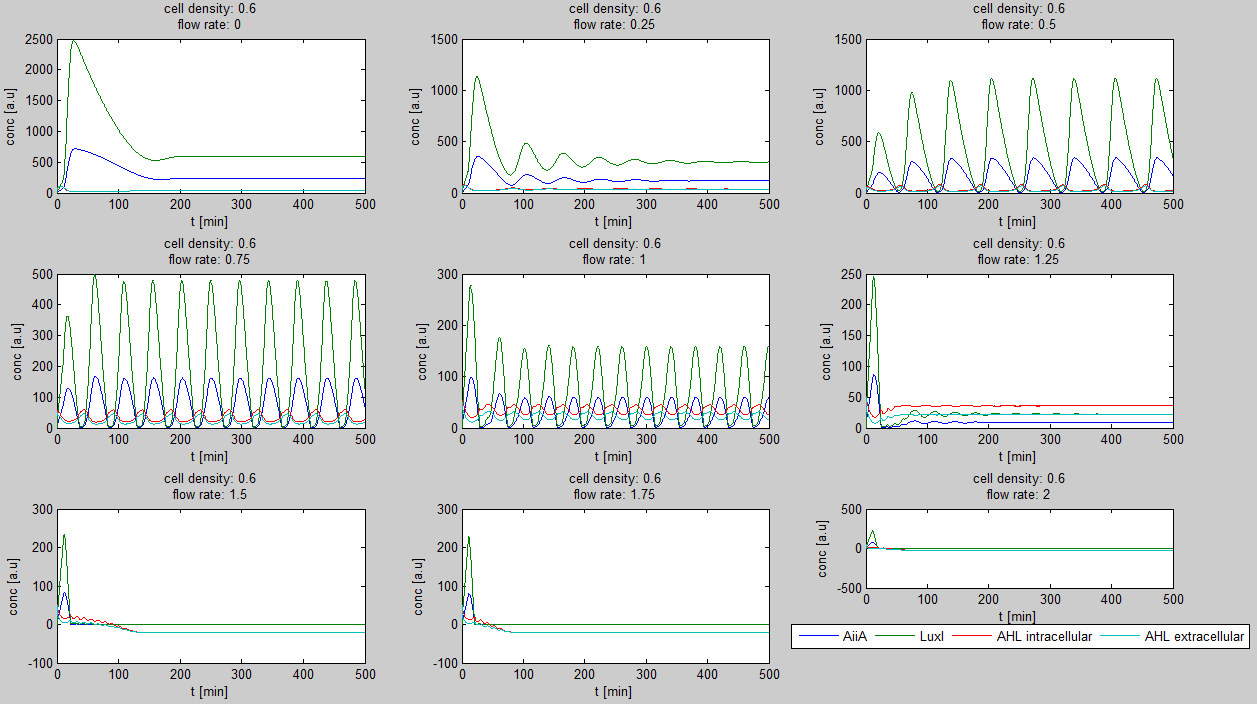
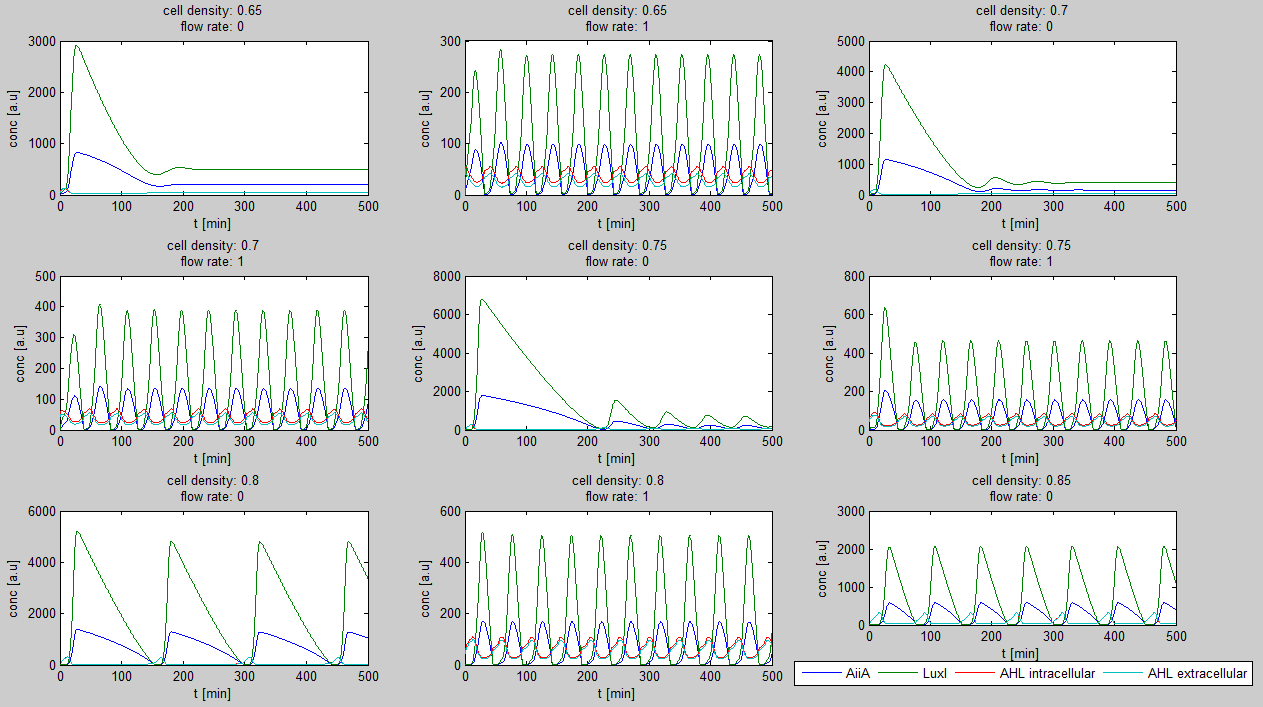
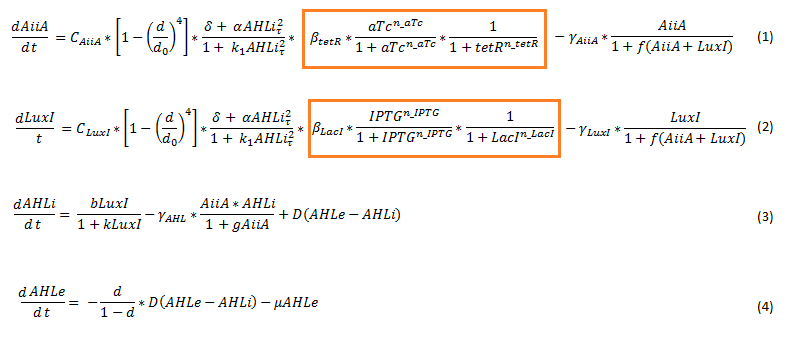
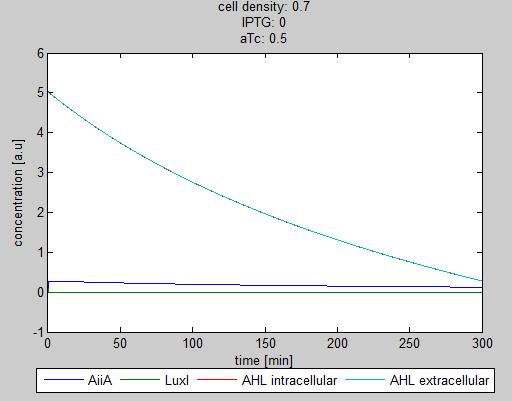
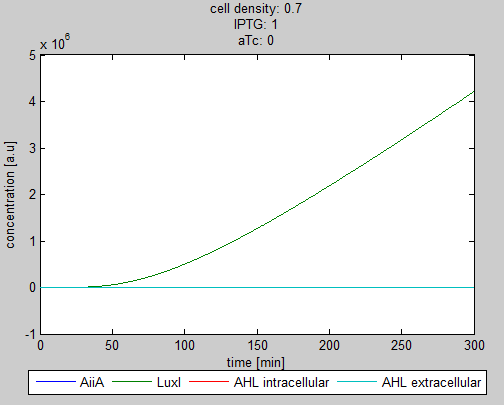
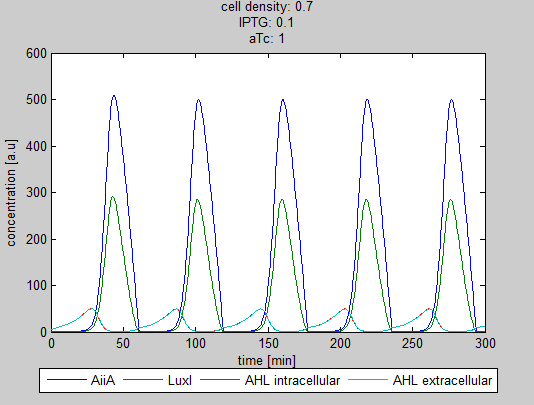
| - | Since our BioBricked oscillatory system is based on the circuit published by Danino et al. in the paper “A synchronized quorum of genetic clocks” [1], our first model of the system is a reproduction of the mathematical model in the supplementary information [2] accompanying the mentioned publication. In their simulations, Danino et al. used a set of four delay differential equations, which we also used as starting point for our modeling work. | + | Since our BioBricked oscillatory system is based on the circuit published by Danino et al. in the paper “A synchronized quorum of genetic clocks” [1], our first model of the system is a reproduction of the mathematical model in the supplementary information [2] accompanying the mentioned publication. In their simulations, Danino et al. used a set of four delay differential equations, which we also used as starting point for our modeling work. |
The steps (transcription, translation, maturation etc.) from the luxI and aiiA genes to the corresponding proteins are not modeled separately. Instead, the delay of the correlation between the internal AHL concentration, which triggers the expression of the genes, and the corresponding AiiA and LuxI concentrations is simulated by a Hill function. This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex. | The steps (transcription, translation, maturation etc.) from the luxI and aiiA genes to the corresponding proteins are not modeled separately. Instead, the delay of the correlation between the internal AHL concentration, which triggers the expression of the genes, and the corresponding AiiA and LuxI concentrations is simulated by a Hill function. This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex. | ||
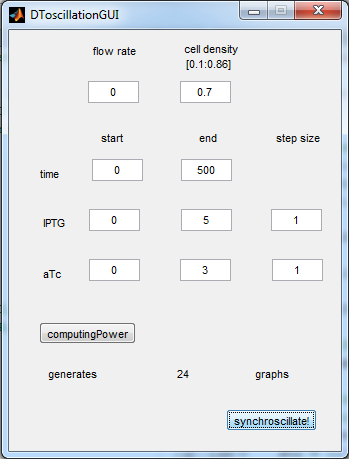
| + | The experimental setup of the cited paper involved the use of microfluidic devices, in which the cell density could be kept constant. Excess cells were flushed away with a predefined flow rate. This also controlled the external AHL concentration. More information can be found in the description about the [[Team:Wageningen_UR/Project/DevicesSetup| setup]] of the device. | ||
| Line 116: | Line 117: | ||
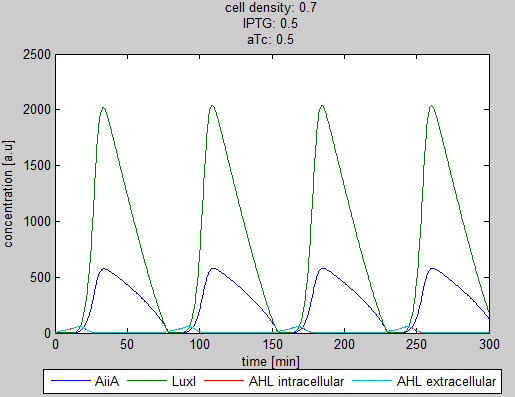
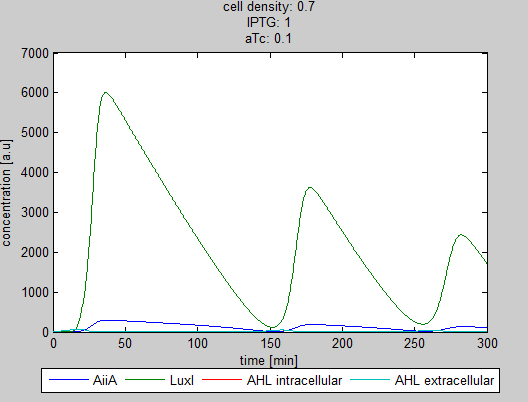
| - | '''Situation 4 and 5:''' ''Interesting behavior of the system when adding IPTG and aTc in reversed proportions. | + | '''Situation 4 and 5:''' ''Interesting behavior of the system when adding IPTG and aTc in reversed proportions.'' |
| + | |||
| + | From the model it can be concluded that the oscillatory behavior can indeed be fine tuned. However, wet-lab experiments are needed to quantify the parameters. | ||
| Line 130: | Line 133: | ||
[2][http://www.nature.com/nature/journal/v463/n7279/suppinfo/nature08753.html Supplementary information] | [2][http://www.nature.com/nature/journal/v463/n7279/suppinfo/nature08753.html Supplementary information] | ||
| - | [3][ | + | [3][https://2007.igem.org/Tokyo/Works Tokyo iGEM 2007] |
}} | }} | ||
Latest revision as of 03:31, 22 September 2011
 "
"