Team:Cambridge/Project/In Vivo
From 2011.igem.org
(→Designing expression constructs) |
|||
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_HEAD}} | ||
| - | = | + | =Expressing reflectin in E. coli= |
| - | + | ||
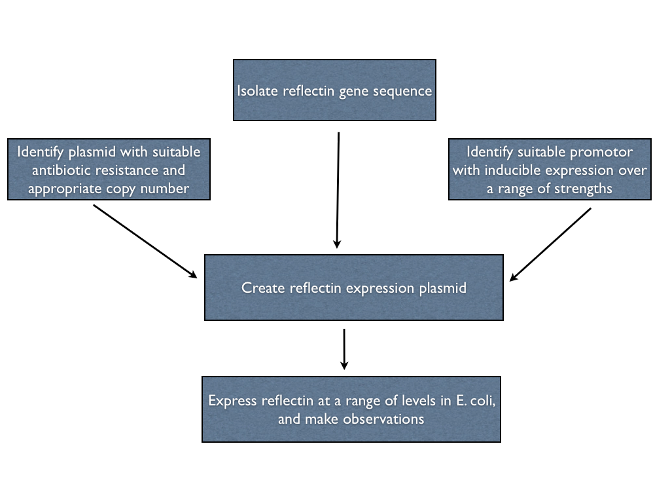
| - | [[File: | + | To help pave the way for the manipulation of living structural colour, we investigated the properties of recombinant reflectins ''in vivo''. We designed multiple expression constructs to allow us to study reflectin under different conditions. An outline of our plan is given below. |
| + | |||
| + | [[File:CAM_invivo_flow.png | centre | 670px]] | ||
==Isolating the reflectin gene== | ==Isolating the reflectin gene== | ||
| - | |||
| - | == | + | We initially attempted to clone reflectin genes from ''Loligo pealeii'' genomic DNA, but this proved a dead end - extracting squid DNA is notoriously difficult and we were unable to obtain samples fresh enough to create a cDNA library. We are very grateful to Wendy Crookes-Goodson for her kind donation of plasmids containing reflectin coding regions codon-optimised for ''E. coli''. |
| + | |||
| + | ==Designing expression constructs== | ||
| + | |||
| + | Expressing recombinant proteins in ''E. coli'' can often lead to misfolding and inclusion body formation, particularly if the proteins are present at concentrations much higher than those a native protein would ever reach. High levels of foreign proteins are likely to disrupt normal cell function, leading to slower growth of cultures or even cell death due to toxic interactions with native ''E. coli'' proteins. As we hoped to observe the effects of [[Team:Cambridge/Project/Background | folded reflectins]] inside living bacterial cells we needed to ensure we limited these problems by reducing the levels of reflectin expression to an acceptable level. | ||
| + | |||
| + | For our ''in vivo'' studies, where correct folding was more important than high yield, we chose the [http://partsregistry.org/Part:pSB3K3 pSB3K3] backbone as it had an origin of replication that would be limited to low-medium numbers of copies per cell. [[Team:Cambridge/Project/In_Vitro | Compare this to our constructs designed for overexpression and purification.]] | ||
| + | |||
| + | Expressing reflectins under an [[Team:Cambridge/Experiments/Low_Level_Expression | arabinose-controlled inducible promoter]] ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I0500 BBa_I0500]) allowed us to titrate expression levels and minimise any toxicity issues that would arise with constitutive expression. | ||
| + | |||
| + | <center> | ||
| + | <gallery widths='300'> | ||
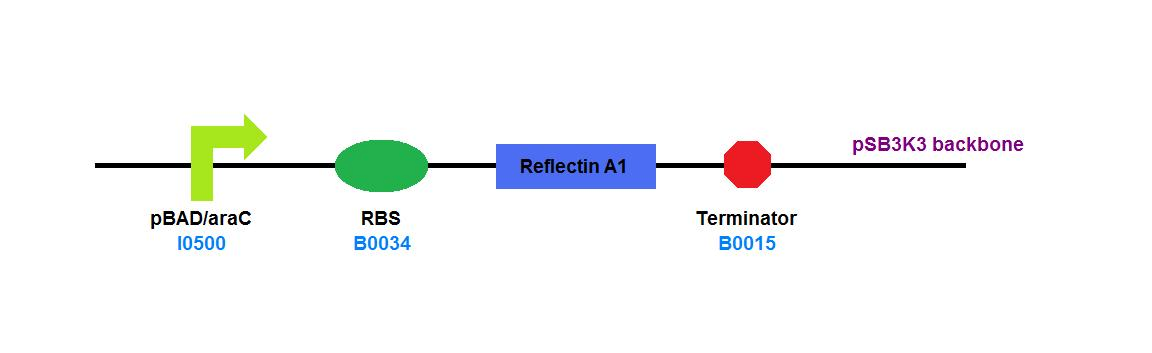
| + | File:Cam_igem_map_GA2.png | [[Team:Cambridge/Experiments/Plasmid_Constructs#GA2 | GA2]] - Reflectin A1 | ||
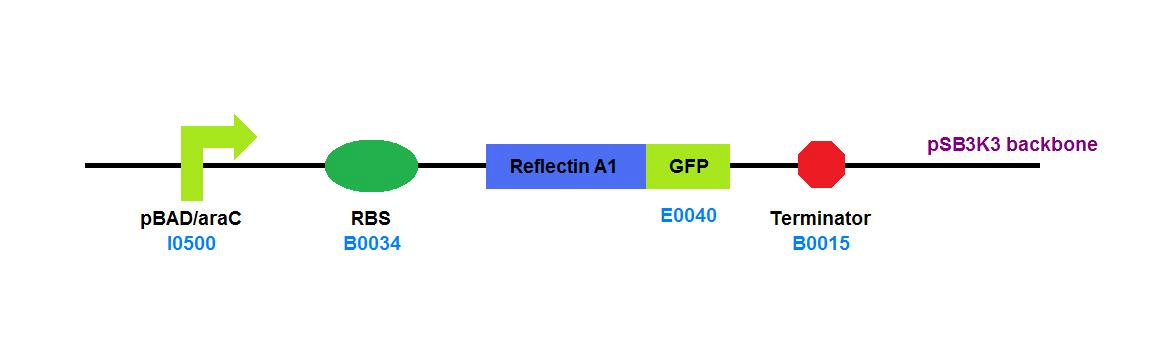
| + | File:Cam_igem_map_GA14.png | [[Team:Cambridge/Experiments/Plasmid_Constructs#GA14 | GA14]] - Reflectin A1 - GFP fusion | ||
| + | </gallery> | ||
| + | </center> | ||
| + | |||
| + | Our experiences with low levels of reflectin expression showed that[[Team:Cambridge/Experiments/Low_Level_Expression | the proteins are soluble and have limited toxicity in ''E. coli'']]. Our reflectin-GFP fusion would not form inclusion bodies, but appeared to be [[Team:Cambridge/Project/Microscopy | uniformly distributed throughout the cell]]. We [[Team:Cambridge/Parts | documented this information]] to aid future research into recombinant reflectins ''in vivo''. | ||
| + | |||
| + | ==Periplasmic Export== | ||
| - | + | [[Team:Cambridge/Project/Background | Reflectins associate with membranes in squid]] to form regular assemblies. In the hope of replicating this behaviour in other cells we attempted to export reflectins into the [http://en.wikipedia.org/wiki/Periplasmic_space periplasmic space] between the two membranes that coat ''E. coli''. | |
| - | + | There are [http://en.wikipedia.org/wiki/Secretion#Secretion_in_Gram_negative_bacteria multiple different export systems] in ''E. coli'' for the secretion of native proteins. As [[Team:Cambridge/Experiments/Low_Level_Expression | reflectins seem to fold into a soluble conformation in the ''E. coli'' cytoplasm]] we chose to harness the Tat pathway as this exports proteins in their fully folded state rather than unfolding them to feed through a channel. | |
| - | + | ||
| - | + | To direct recombinant proteins into the periplasm we needed to create fusion proteins between reflectins and the N terminal region of normally secreted proteins, to allow recognition by the export machinery. Our first choice was an export sequence from the membrane protein TorA, which had previously been used to [http://www.ncbi.nlm.nih.gov/pubmed/11123687 export correctly folded GFP into the periplasm]. This allowed us to use GFP fusions as an easy to observe reporter of successful export. | |
| - | + | ||
| + | Unfortunately, as of the time of writing we have not observed successful export of reflectin-GFP fusions. [[Team:Cambridge/Experiments/Periplasmic_Export | Read more about our export system here.]] | ||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | ||
Latest revision as of 14:52, 21 September 2011
Contents |
Expressing reflectin in E. coli
To help pave the way for the manipulation of living structural colour, we investigated the properties of recombinant reflectins in vivo. We designed multiple expression constructs to allow us to study reflectin under different conditions. An outline of our plan is given below.
Isolating the reflectin gene
We initially attempted to clone reflectin genes from Loligo pealeii genomic DNA, but this proved a dead end - extracting squid DNA is notoriously difficult and we were unable to obtain samples fresh enough to create a cDNA library. We are very grateful to Wendy Crookes-Goodson for her kind donation of plasmids containing reflectin coding regions codon-optimised for E. coli.
Designing expression constructs
Expressing recombinant proteins in E. coli can often lead to misfolding and inclusion body formation, particularly if the proteins are present at concentrations much higher than those a native protein would ever reach. High levels of foreign proteins are likely to disrupt normal cell function, leading to slower growth of cultures or even cell death due to toxic interactions with native E. coli proteins. As we hoped to observe the effects of folded reflectins inside living bacterial cells we needed to ensure we limited these problems by reducing the levels of reflectin expression to an acceptable level.
For our in vivo studies, where correct folding was more important than high yield, we chose the [http://partsregistry.org/Part:pSB3K3 pSB3K3] backbone as it had an origin of replication that would be limited to low-medium numbers of copies per cell. Compare this to our constructs designed for overexpression and purification.
Expressing reflectins under an arabinose-controlled inducible promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I0500 BBa_I0500]) allowed us to titrate expression levels and minimise any toxicity issues that would arise with constitutive expression.
GA2 - Reflectin A1 |
GA14 - Reflectin A1 - GFP fusion |
Our experiences with low levels of reflectin expression showed that the proteins are soluble and have limited toxicity in E. coli. Our reflectin-GFP fusion would not form inclusion bodies, but appeared to be uniformly distributed throughout the cell. We documented this information to aid future research into recombinant reflectins in vivo.
Periplasmic Export
Reflectins associate with membranes in squid to form regular assemblies. In the hope of replicating this behaviour in other cells we attempted to export reflectins into the [http://en.wikipedia.org/wiki/Periplasmic_space periplasmic space] between the two membranes that coat E. coli.
There are [http://en.wikipedia.org/wiki/Secretion#Secretion_in_Gram_negative_bacteria multiple different export systems] in E. coli for the secretion of native proteins. As reflectins seem to fold into a soluble conformation in the E. coli cytoplasm we chose to harness the Tat pathway as this exports proteins in their fully folded state rather than unfolding them to feed through a channel.
To direct recombinant proteins into the periplasm we needed to create fusion proteins between reflectins and the N terminal region of normally secreted proteins, to allow recognition by the export machinery. Our first choice was an export sequence from the membrane protein TorA, which had previously been used to [http://www.ncbi.nlm.nih.gov/pubmed/11123687 export correctly folded GFP into the periplasm]. This allowed us to use GFP fusions as an easy to observe reporter of successful export.
Unfortunately, as of the time of writing we have not observed successful export of reflectin-GFP fusions. Read more about our export system here.
 "
"