Team:Harvard/Template:NotebookDataAugust
From 2011.igem.org
(Difference between revisions)
(Created page with "<div> ==August 1st== ===Team Wolfe=== '''Optimizing PCR reaction for pSB4K5:''' The gradient PCR did work, but the yield was still very low. We will try a few different approach...") |
|||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <div> | + | <div id='801' style="display:none"> |
==August 1st== | ==August 1st== | ||
===Team Wolfe=== | ===Team Wolfe=== | ||
| Line 10: | Line 10: | ||
*Gel stab went into 20µL H2O; 66 C annealing, 3 min elongation; pSB4K5_F+ZFBhl and pSB4K5_R+URA3hl primers | *Gel stab went into 20µL H2O; 66 C annealing, 3 min elongation; pSB4K5_F+ZFBhl and pSB4K5_R+URA3hl primers | ||
*Results: unsuccessful. Perhaps the gel stab did not give an adequate amount of template. | *Results: unsuccessful. Perhaps the gel stab did not give an adequate amount of template. | ||
| - | [[File: | + | [[File: HARV2011.08.01. pSB4K5 grad stab(labeled).png|thumb|left|7/29 pSB4K5 gradient samples for gel stab]] |
| - | [[File: | + | [[File: HARV2011.08.01.pSB4K5stab_zeorpoZ_ZF(labeled).png|thumb|none|pSB4K5 gel stab PCR results 8/1/11]] |
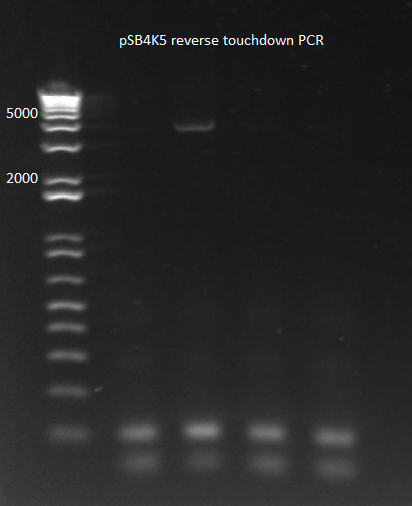
2) Reverse Touchdown PCR: | 2) Reverse Touchdown PCR: | ||
| Line 18: | Line 18: | ||
*3 min elongation | *3 min elongation | ||
*Results: one of the four reactions worked. This is very strange since the reactions should have been identical. | *Results: one of the four reactions worked. This is very strange since the reactions should have been identical. | ||
| - | [[File: | + | [[File: HARV2011.08.01.pSB4K5 reverse touchdown(labeled).png|thumb|none|pSB4K5 reverse touchdown 8/1/11]] |
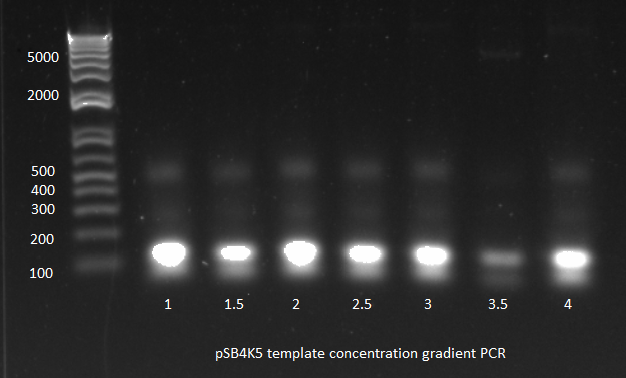
3) Template concentration gradient PCR: | 3) Template concentration gradient PCR: | ||
| Line 25: | Line 25: | ||
*66 C anneal, 3 min elongation | *66 C anneal, 3 min elongation | ||
*Results: only one of the reactions worked. We did use the gradient cycler which has not always worked well for us. We also noticed that when we removed the tubes from the cycler, most of the liquid had condensed on the lids. | *Results: only one of the reactions worked. We did use the gradient cycler which has not always worked well for us. We also noticed that when we removed the tubes from the cycler, most of the liquid had condensed on the lids. | ||
| - | [[File: | + | [[File: HARV2011.08.01.pSB4K5 template conc grad(labeled).png|thumb|none|pSB4K5 template concentration gradient PCR 8/1/11]] |
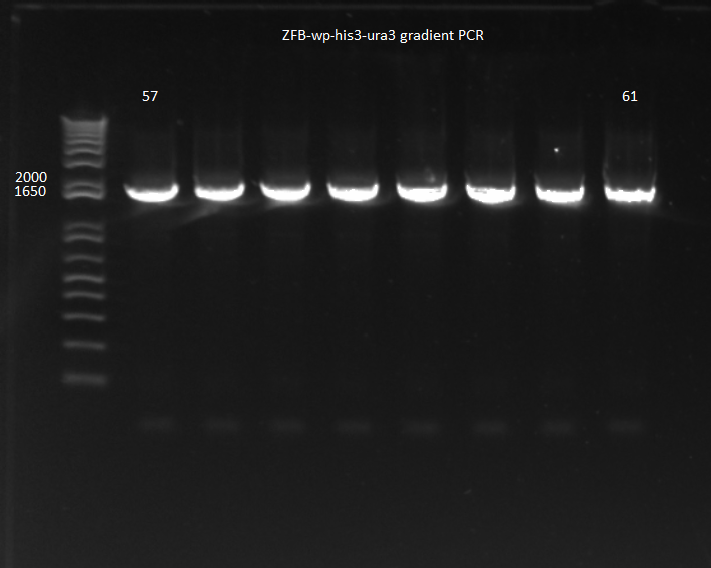
4) Gradient PCR (overnight): | 4) Gradient PCR (overnight): | ||
| Line 36: | Line 36: | ||
*because of a substantial difference in primer melting temperatures, we did a 57-61 C annealing gradient; 90 sec elongation | *because of a substantial difference in primer melting temperatures, we did a 57-61 C annealing gradient; 90 sec elongation | ||
*Results: all 8 conditions worked! | *Results: all 8 conditions worked! | ||
| - | [[File: | + | [[File: HARV2011.08.01.ZFBhisura(labeled).png|thumb|none|ZFB-wp-his3-ura3 gradient PCR 8/1/11]] |
*Minelute PCR purification of 4 of the reactions: yield=epic. | *Minelute PCR purification of 4 of the reactions: yield=epic. | ||
| - | [[File: | + | [[File: HARVEpic iGEM screen capture.jpeg|thumb|none|BEST YIELD EVER!]] |
'''pSR01 plate reading:''' | '''pSR01 plate reading:''' | ||
| Line 68: | Line 68: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.1_cjun_restriction_digest_annotated.png|thumb|We can clearly see the appropriate sized bands (717 and 578 bps) after cutting our cross junction product with Bsa1]] |
|} | |} | ||
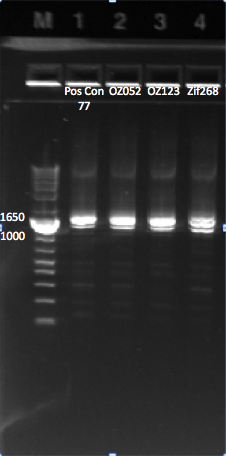
====Redoing the isothermal assembly/transformation for Pos con 77 and the 3 controls==== | ====Redoing the isothermal assembly/transformation for Pos con 77 and the 3 controls==== | ||
| - | Today we redid the isothermal assembly for Pos con 77, Zif268, OZ052, and OZ123 using our | + | Today we redid the isothermal assembly for Pos con 77, Zif268, OZ052, and OZ123 using our previous protocol. We also performed a chemical transformation using Top 10 ChemComp cells, and plated them with 100 ul each plate. We will have to wait and see whether colonies grow tomorrow. |
| - | We also performed a cross junction PCR on the isothermal assembly product to make sure that it worked, using our | + | We also performed a cross junction PCR on the isothermal assembly product to make sure that it worked, using our usual protocol. The gel (pictured below) shows appropriately sized bands for the cross junction. |
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.1_pos_con_77_f2f3_controls_annotated.png|thumb|All our isothermal assemblies worked, since we can see bands at ~1.4 kb.]] |
|} | |} | ||
| Line 90: | Line 90: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.1_cb_bottom_plamid_digestion_annotated.png|thumb|Product after digesting our plasmid with Bsa1]] |
|} | |} | ||
| Line 98: | Line 98: | ||
**Performed PCR and ran gel on products | **Performed PCR and ran gel on products | ||
***Successful!! | ***Successful!! | ||
| - | [[File: | + | [[File: HARV2011.08.01.Zeocincheckfroplatereader(labeled).png|thumb|none|8/1/11 Zeocin insertion into ECNR2 for plate reader]] |
====Transformation of Zif268 into ECNR2+zeocin without Kan+ZFB+wp==== | ====Transformation of Zif268 into ECNR2+zeocin without Kan+ZFB+wp==== | ||
| Line 115: | Line 115: | ||
***This served as a postive control | ***This served as a postive control | ||
***Grown in .005% SDS and 2,4,8,12,16 µM of IPTG | ***Grown in .005% SDS and 2,4,8,12,16 µM of IPTG | ||
| - | *Plate was taken down to plate reader at 5:25 pm and began a 16 hour program | + | *Plate was taken down to plate reader at 5:25 pm and began a 16 hour program</div> |
| - | + | <div id='802' style="display:none"> | |
==August 2nd== | ==August 2nd== | ||
===Team TolC=== | ===Team TolC=== | ||
| Line 128: | Line 128: | ||
{| | {| | ||
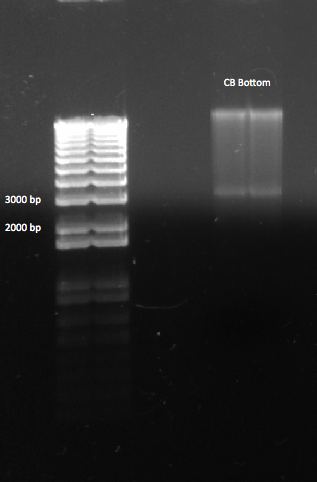
| - | |[[File: | + | |[[File:HARV2011.8.2_miniprep_comparison.jpg|thumb|Miniprep results. PZE22G had excellent yields, while CB Bottom did not.]] |
|} | |} | ||
| Line 150: | Line 150: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.2_oligo_digestion_1_annotated.png|thumb|Our first digestion did not show any bands below 100 bp.]] |
|} | |} | ||
| Line 162: | Line 162: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.2_oligo_digestion_2_annotated.png|thumb|The bands on this 1.2% e-gel were unexpectedly high. It maybe a problem with the gel.]] |
|} | |} | ||
| Line 173: | Line 173: | ||
In order to further troubleshoot our less-than-ideal gels for our digestion product, we ran a third digestion reaction, using the second recipe. This time we simultaneously digested our oligo, and our cross junction (which had been shown to work before). We also ran the unpurified PCR product from recipe 2 on this gel. | In order to further troubleshoot our less-than-ideal gels for our digestion product, we ran a third digestion reaction, using the second recipe. This time we simultaneously digested our oligo, and our cross junction (which had been shown to work before). We also ran the unpurified PCR product from recipe 2 on this gel. | ||
| - | See | + | See tomorrow's entry for the gel image. |
Our digestion of the cross junction appeared to have worked, while once again, we got unusually large bands for our oligo. | Our digestion of the cross junction appeared to have worked, while once again, we got unusually large bands for our oligo. | ||
| Line 179: | Line 179: | ||
We performed a PCR to clone out Zif268 so that we can later assemble Zif and GFP into our reporter plasmid. We used the Zif clone Forward and Zif clone Reverse primers (Primer index: 148 and 149). We had an annealing temperature of 62* and an extension time of 90 seconds. Our expected product was ~3 kb. | We performed a PCR to clone out Zif268 so that we can later assemble Zif and GFP into our reporter plasmid. We used the Zif clone Forward and Zif clone Reverse primers (Primer index: 148 and 149). We had an annealing temperature of 62* and an extension time of 90 seconds. Our expected product was ~3 kb. | ||
| - | See | + | See tomorrow's entry for the gel. Our PCR appeared to have worked, and we have a strong band at our expected product size. |
===Team Wolfe=== | ===Team Wolfe=== | ||
| Line 185: | Line 185: | ||
*We got some beautiful growth curves! A few of the more critical ones are below: | *We got some beautiful growth curves! A few of the more critical ones are below: | ||
*NM+his: both selection strain and pSR01 grew, but the selection strain grew faster. This might just be an artifact from having accidentally added more cells to the selection strain than the pSR01 wells, but it may also show that the pSR01 strain grows slower. Another possibility is that we are expressing enough zinc fingers for it to be slightly toxic for the cells. Next time maybe we should let the plate reader go for longer than 16 hours just in case the cells need more time. | *NM+his: both selection strain and pSR01 grew, but the selection strain grew faster. This might just be an artifact from having accidentally added more cells to the selection strain than the pSR01 wells, but it may also show that the pSR01 strain grows slower. Another possibility is that we are expressing enough zinc fingers for it to be slightly toxic for the cells. Next time maybe we should let the plate reader go for longer than 16 hours just in case the cells need more time. | ||
| - | [[File: | + | [[File: HARV2011.08.01.NM+his.png|none|NM+his: 162 and 172 selection strain, 182 and 192 pSR01]] |
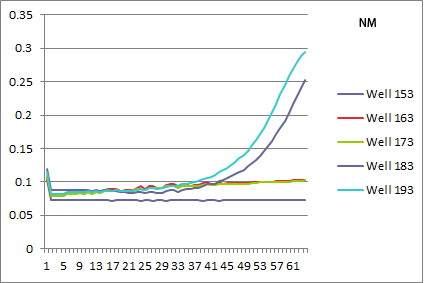
*NM: selection strain does not grow, while pSR01 does a perfect rescue! | *NM: selection strain does not grow, while pSR01 does a perfect rescue! | ||
| - | [[File: | + | [[File: HARV2011.08.01.NM.png|none|NM: 163 and 173 selection strain, 183 and 193 pSR01]] |
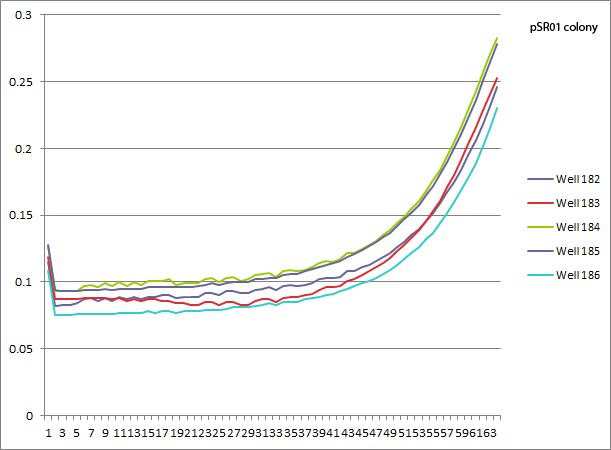
| - | *pSR01 | + | *pSR01 HARV3-AT comparison: the concentration of 3-AT does not seem to make a difference in the growth curves (the differences appear to be due to different wells starting with slightly different amounts of cells). Next time we can try a higher 3-AT concentration, but since we expect Zif268 to bind strongly this might just be what you get. |
| - | [[File: | + | [[File: HARV2011.08.01.pSR01 comparison.png|none|pSR01 colony comparison: 182 NM+his, 183 NM, 184 1mM 3-AT, 185 2.5mM 3-AT, 186 5mM 3-AT]] |
*NM+his+IPTG: IPTG more or less killed the pSR01 cells but not the selection strain. This may be caused by too many zinc fingers being expressed, so we may want to try less IPTG. | *NM+his+IPTG: IPTG more or less killed the pSR01 cells but not the selection strain. This may be caused by too many zinc fingers being expressed, so we may want to try less IPTG. | ||
| - | [[File: | + | [[File: HARV2011.08.01.NM+his+IPTG.png|none|NM+his+IPTG: 167 and 177 selection strain, 187 and 197 pSR01]] |
'''EcNR2 Wolfe Selection system: MAGE''' | '''EcNR2 Wolfe Selection system: MAGE''' | ||
| Line 199: | Line 199: | ||
'''pSB4K5:''' | '''pSB4K5:''' | ||
*Our pSB4K5 PCR still eludes us. Our overnight gradient did work, but no better than the other days' reactions. | *Our pSB4K5 PCR still eludes us. Our overnight gradient did work, but no better than the other days' reactions. | ||
| - | [[File: | + | [[File: HARV2011.08.02.pSB4K5 gradient pcr1(labeled).png|thumb|none|pSB4K5 gradient 8/2/11]] |
| - | *Gel extraction and Minelute only gave us a 9.4ng/µL yield (260/280=1.82) which is not enough for isothermal assembly. We will have to miniprep more plasmid for template for PCRs, and we might want to consider using ligation or some other method. | + | *Gel extraction and Minelute only gave us a 9.4ng/µL yield (260/280=1.82) which is not enough for isothermal assembly. We will have to miniprep more plasmid for template for PCRs, and we might want to consider using ligation or some other method.</div> |
| - | + | <div id='803' style="display:none"> | |
==August 3rd== | ==August 3rd== | ||
===Team ZF=== | ===Team ZF=== | ||
| Line 207: | Line 207: | ||
We ran a gel of our digestion product from yesterday, and our PCR to clone out Zif 268 to construct the reporter plasmid. | We ran a gel of our digestion product from yesterday, and our PCR to clone out Zif 268 to construct the reporter plasmid. | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.8_digest_PCR_annotated.png|thumb|Our results for the final digestion we ran yesterday, and the PCR to clone out Zif 268.]] |
|} | |} | ||
| Line 217: | Line 217: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.3_pc77_swap_minipreps.jpg|thumb|Results of our miniprep.]] |
|} | |} | ||
Once again, we got unusually low yields. We decided to test the quality of our spec, to see if that was the problem. We grew PZE22G(which does not have spec resistance) in spec to see if it grew. By the end of the day, nothing had grown. | Once again, we got unusually low yields. We decided to test the quality of our spec, to see if that was the problem. We grew PZE22G(which does not have spec resistance) in spec to see if it grew. By the end of the day, nothing had grown. | ||
| - | Still, we decided to PCR the miniprep product, since any DNA that is in the product should be amplified. We performed our | + | Still, we decided to PCR the miniprep product, since any DNA that is in the product should be amplified. We performed our usual protocol for a cross junction PCR, but we repeated the reaction for 30 cycles. |
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.3_junction_pcr_annotated.png|thumb]] |
| - | |[[File: | + | |[[File:HARV2011.8.3_junction_pcr_2_and_gfp_annotated.png|thumb]] |
|} | |} | ||
| Line 234: | Line 234: | ||
'''PCRs of Zif 268 and GFP''' | '''PCRs of Zif 268 and GFP''' | ||
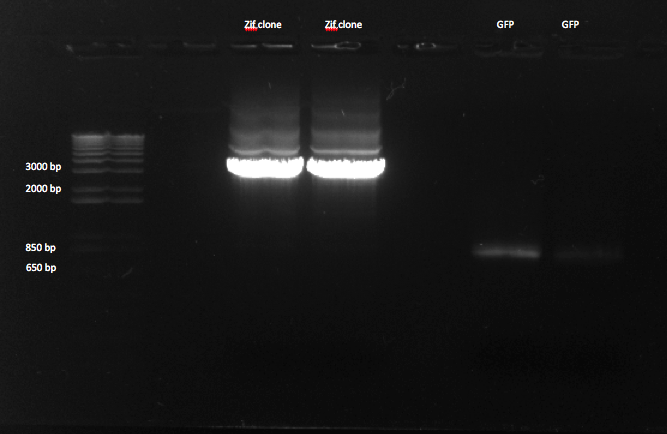
| - | We decided to perform more PCRs to clone out Zif 268, since it will have to be gel purified. This reaction had the same conditions as | + | We decided to perform more PCRs to clone out Zif 268, since it will have to be gel purified. This reaction had the same conditions as yesterday's. |
We also received the GFP plate today, so we decided to perform a colony PCR in order to get out the sequence that encodes GFP. We used our GFP Forward and GFP Reverse (Primer Index: 146 and 147) for the reaction, had an annealing temperature of 58*, and an extension time of 30 seconds. The expected product size was ~700 bp. | We also received the GFP plate today, so we decided to perform a colony PCR in order to get out the sequence that encodes GFP. We used our GFP Forward and GFP Reverse (Primer Index: 146 and 147) for the reaction, had an annealing temperature of 58*, and an extension time of 30 seconds. The expected product size was ~700 bp. | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.3_junction_pcr_2_and_gfp_annotated.png|thumb|The last four lanes show the expected band size of ~825 bp for GFP. However the bands are comparatively faint.]] |
|} | |} | ||
| Line 247: | Line 247: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.3_zif_clone_and_gfp_annotated.png|thumb|We made a 1% agarose gel to gel purify our zif clone and GFP. The bands for GFP are much fainter than those for Zif 268.]] |
|} | |} | ||
| Line 255: | Line 255: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.8.3_gfp_for_gel_extr._annotated.png|thumb|We ran another PCR to get out GFP.]] |
|} | |} | ||
| Line 282: | Line 282: | ||
{| | {| | ||
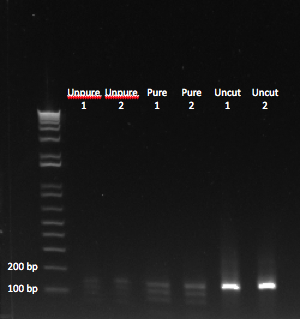
| - | |[[File: | + | |[[File:HARV2011.8.3_double_digested_annotated.png|thumb|We could see bands of the appropriate size, after digesting our PCR product. Unpure = cut without PCR purification.]] |
|} | |} | ||
| Line 312: | Line 312: | ||
**PCR failed | **PCR failed | ||
| - | [[File: | + | [[File:HARV2011.08.03.pSB4K5 rtd-0012.png|thumb|none|A successful PCR would have resulted in a 3.5-4Kb band.]] |
*Miniprep of pSB4K5 strain for future PCR yielded 15ng/µL | *Miniprep of pSB4K5 strain for future PCR yielded 15ng/µL | ||
| Line 331: | Line 331: | ||
===Team TolC=== | ===Team TolC=== | ||
*Set up and ran plate reader for the growth rates of ECNR2+kan-ZFB-wp+zeocin+spec plasmid, ECNR2+kan-ZFB-wp+zeocin Δspec plasmid, ECNR2, and ECNR2+zeocin+spec plasmid Δkan-ZFB-wp in .04% SDS and 25, 12.5, 6.25, 3.125, 1.5625 µM</div> | *Set up and ran plate reader for the growth rates of ECNR2+kan-ZFB-wp+zeocin+spec plasmid, ECNR2+kan-ZFB-wp+zeocin Δspec plasmid, ECNR2, and ECNR2+zeocin+spec plasmid Δkan-ZFB-wp in .04% SDS and 25, 12.5, 6.25, 3.125, 1.5625 µM</div> | ||
| + | <div id='804' style="display:none"> | ||
| + | ==August 4th== | ||
| + | ===Team ZF=== | ||
| + | ====Pos Con 77 and control swap sequencing results==== | ||
| + | {| | ||
| + | | align="center" style="background:#f0f0f0;"|''' ''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Pos. Con. 77''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Notes''' | ||
| + | |- | ||
| + | | ||1||lots of snps and deletions | ||
| + | |- | ||
| + | | ✓||2||G->C | ||
| + | |- | ||
| + | | ||3||deletion before type II cut site | ||
| + | |- | ||
| + | | ||4||deletion in terminator | ||
| + | |- | ||
| + | | align="center" style="background:#f0f0f0;"|''' ''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''OZ052 swap''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Notes''' | ||
| + | |- | ||
| + | | ✓||1||G->C | ||
| + | |- | ||
| + | | ✓||2||G->C | ||
| + | |- | ||
| + | | ||3||deletions and mismatches in ultramer | ||
| + | |- | ||
| + | | ||4||terrible | ||
| + | |- | ||
| + | |||
| + | |||
| + | | align="center" style="background:#f0f0f0;"|''' ''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''OZ123 swap''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Notes''' | ||
| + | |- | ||
| + | | ✓||1||fine, deletion at end | ||
| + | |- | ||
| + | | ||2||deletions and snps | ||
| + | |- | ||
| + | | ||3||long deletion in helix | ||
| + | |- | ||
| + | | ✓||4||fine | ||
| + | |- | ||
| + | | align="center" style="background:#f0f0f0;"|''' ''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Zif268 swap''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Notes''' | ||
| + | |- | ||
| + | | ✓||1||fine, TGEKP instead of TGQKP | ||
| + | |- | ||
| + | | ✓||2||G->C, TGEKP instead of TGQKP | ||
| + | |- | ||
| + | | ||3||terrible | ||
| + | |- | ||
| + | | ✓||4||G->C, TGEKP instead of TGQKP | ||
| + | |} | ||
| + | |||
| + | We had a good sequence for all of our samples. We made 2 ml cultures of these, to miniprep, make final plates, and glycerol stocks. Note, that for Zif 268, the linker between F1/F2 is 'TGEKP', as that was the sequence in the ultramer we ordered. The actual sequence is 'TGQKP'. If this proves to be a problem, we can always try using quick change to insert the appropriate codon. | ||
| + | |||
| + | ====Checking on the reporter strain==== | ||
| + | Our transformed bacteria with the 11.3 ul isothermal assembly product and 100 ng backbone grew. We picked four from the plate and are growing 2 ml cultures from time to miniprep and send for sequencing tomorrow. | ||
| + | |||
| + | Our other bacteria with 10 ul product, but less than 100 ng backbone, did not grow. | ||
| + | |||
| + | ====Ligation of oligos and backbone==== | ||
| + | '''Checking our plates''' | ||
| + | |||
| + | No colonies grew on any of our plates that contained bacteria transformed with ligation product. We wondered if this was due to our ligase no longer working, so we decided to test our ligase. | ||
| + | |||
| + | '''Testing our ligase''' | ||
| + | |||
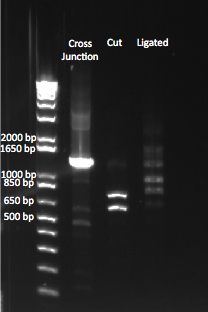
| + | We wanted to test our ligase by cutting our cross junction with Xba1, and then ligating it, so see if it formed the original cross junction. We ran a gel comparing our whole cross junction, our digested product, and our ligated product. | ||
| + | |||
| + | {| | ||
| + | |[[File:HARV2011.8.4_xba1_digest_and_ligation_annotated.png|thumb|Comparing our original, cut, and ligated cross junction.]] | ||
| + | |} | ||
| + | |||
| + | Our uncut cross junction was ~1.4 kb. We saw the appropriate sized bands of 593 and 739 bp after cutting the junction with Xba1. However, after ligation, we saw bands of varying lengths, at the one we expected (at ~1.4 kb) was not noticeably brighter than the other bands. We are unsure what these bands are. | ||
| + | |||
| + | '''Cutting and gel purifying CB Bottom''' | ||
| + | |||
| + | We also wondered if our unsuccessful ligation was due to an ineffective CB Bottom segment. We had been unsure exactly which band was the appropriate one, and so we cut both. Just in case we decided to re-do the digestion of the CB Bottom plasmid, with Bsa1. We used the following recipe: | ||
| + | *46 ul DNA (to get 1 ug) | ||
| + | *1 ul enzyme | ||
| + | *5 ul Buffer T4 | ||
| + | *0.6 ul BSA | ||
| + | |||
| + | We then ran the product on a 0.7% agarose gel: | ||
| + | |||
| + | {| | ||
| + | |[[File:HARV2011.8.4_cb_bottom_for_extraction_annotated.png|thumb|A gel of the CB Bottom backbone, after digestion. We expected a band at ~2.7 kb]] | ||
| + | |} | ||
| + | |||
| + | We cut out the appropriate band, and put it in the fridge to gel purify tomorrow. | ||
| + | |||
| + | ====Making more Zif268 (whole) plasmid==== | ||
| + | We performed a miniprep of the Zif268 culture we grew yesterday. The miniprep had a yield of ~16 ng/ul. | ||
| + | |||
| + | ===Team Wolfe=== | ||
| + | '''Plate Reader''' | ||
| + | *Looks great! | ||
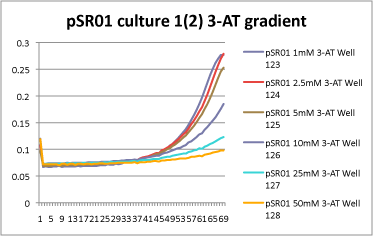
| + | *1mM, 2.5mM, and 5mM 3-AT have slight differences in growth rate, but it is hard to tell how significant they are. However, starting with 10mM 3-AT, the growth rate drops dramatically! Here is an example from 1 culture set: | ||
| + | [[File: HARV2011.08.03.pSR01 1(2) 3-AT gradient-0013.png|none|pSR01 culture 1 (duplicate 2) 3-AT gradient]] | ||
| + | |||
| + | '''MAGE 3 HisB and PyrF Knock Out Gel''' | ||
| + | *HisB Check | ||
| + | ** Control reaction showed a nice strong band. All the other lanes had bands, but they varied in strength. It looks like we may have some hits, but we need our WT control primers to come in for us to be sure. | ||
| + | *PyrF check | ||
| + | **Similar to HisB--all the lanes had bands but some were brighter than others. | ||
| + | |||
| + | [[File:HARV2011.08.04 HisBUra3 MAGE 3 Deletion Check (labeled)-0014.png|thumb|none|HisB]] | ||
| + | [[File:HARV2011.08.04 PyrF MAGE 3 Deletion Check 2s exposure (labeled)-0015.png|thumb|none|PyrF]] | ||
| + | |||
| + | '''ZFB-wp-his3-ura3 restriction enzyme PCR''' | ||
| + | *PCR to add Spe1 and Xba1 sites to join insert with pSB4K5 plasmid | ||
| + | *Spe1 ZFB_F and Xba1 URA3-R primers | ||
| + | *90 sec elongation, 62˚C annealing | ||
| + | |||
| + | ===Team TolC=== | ||
| + | ====Plate Reader==== | ||
| + | {| | ||
| + | |[[File:HARV2011.8.4graphofKNplatereader.png|thumb|Plate Reader Graph of KN.]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
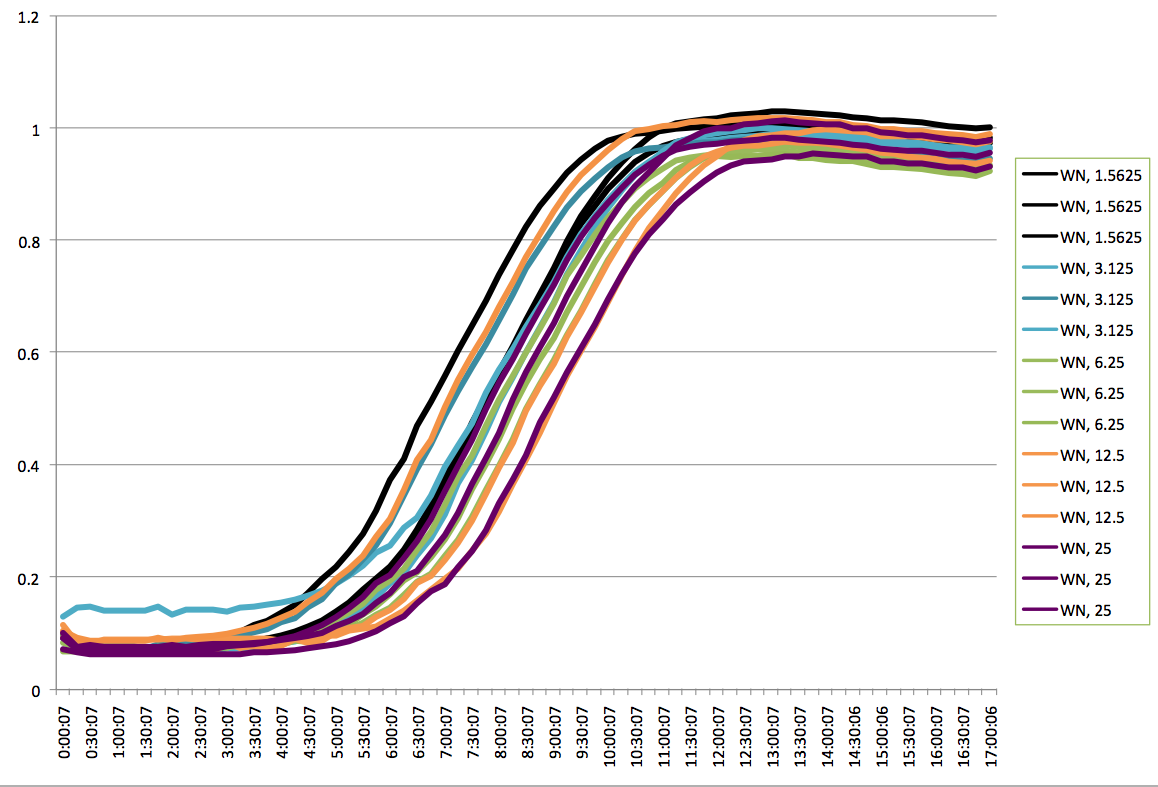
| + | |[[File:HARV2011.8.4graphofWNplatereader.png|thumb|Plate Reader Graph of WN.]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
| + | |[[File:HARV2011.8.4graphofKSplatereader.png|thumb|Plate Reader Graph of KS.]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
| + | |[[File:HARV2011.8.4graphofWSplatereader.png|thumb|Plate Reader Graph of WS.]] | ||
| + | |} | ||
| + | |||
| + | ====PCR of plate reader colonies==== | ||
| + | *Made glycerol stock of ECNR2+spec plasmid+zeocin, WS</div> | ||
 "
"