Team:Freiburg/Description
From 2011.igem.org
(Difference between revisions)
(→Part design) |
(→Precipitator) |
||
| Line 34: | Line 34: | ||
We only submitted one of the three versions, to reduce redundancy in the registry. Please contact us for any questions. | We only submitted one of the three versions, to reduce redundancy in the registry. Please contact us for any questions. | ||
| + | |||
| + | ===Plastic binding domain=== | ||
| + | |||
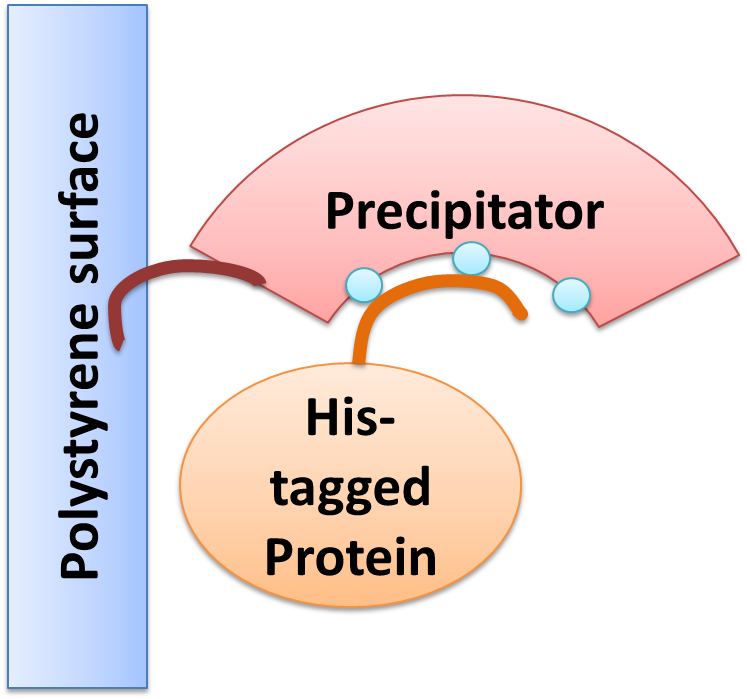
| + | One oft the issues of our project „lab in a cell“ was to use endogenous proteins produced by the cell itself for specific purification and hereby to replace expensive columns. The „precipitator“ designed by our team contains a protein binding domain which complexes nickel and thus enables the binding of His-tagged proteins. After cell lysis the “precipitator” is freely dissolved in the cell lysate. To be able to insolate the “precipitator”-His-tagged-protein-complex from the other cell components it has to be immobilized by another protein domain. | ||
| + | |||
| + | During routine phage display of random peptide libraries, phages were found that bound directly to the plastic surface of the used plastic micro titer plates. The number of plastic binding phages obtained during the phage display experiments depended on the saturation of the plastic micro titer plates with target protein for the antibody-binding phages and could be reduced by the use of blocking proteins as BSA or non-fat milk. Plastic binding phages were resistant to washing steps with PBS alone as well as to PBS in combination with BSA or non-fat dry milk. It was shown that plastic binding phages were even more difficult to recover by acid elution than the “normal” antibody binding phages (Adey et al., 1995). | ||
| + | |||
| + | The binding strength of the plastic binding protein was best on polystyrene plastic surfaces and also observed on PVC-plates (Adey et al., 1995). | ||
| + | |||
| + | The mechanisms by which the phage surface proteins bind to plastic are not well understood. The plastic binding amino acid sequences showed no obvious sequence similarity but where generally enriched by Tyr and Trp residues and are completely devoid of Cys residues. It is possible that the binding comes off non-specific hydrophobic interactions due to partial denaturation of the protein (Cantanero et al., 1980) and potentially due to interactions of the Tyr and Trp residues with the aromatic moieties of the polystyrene plastic surface of micro titer plates. | ||
| + | |||
| + | As well as high hydrophobicity does not necessarily imply plastic binding quality (Menendez et al., 2005) the observed plastic binding phages showed hydrophobic peptide sequences on their surfaces. Expressing such hydrophobic proteins in our host organism E.coli can lead to problems with inclusion bodies or decreased vitality up to cell death. | ||
| + | |||
| + | During our experiments we couldn’t obtain any clones containing both the gene for the plastic binding protein and a constitutive promoter-RBS-construct. We finally succeeded in expressing the plastic binding domain using an inducible promoter. To test the plastic binding domain and get data for our modeling we hereby used an IPTG-inducible promoter. In our completed “Lab in a cell” model the plastic binding domain, as a part of the “precipitator” would be expressed by one of our light inducible promoters. | ||
==<span style="color:green;">Green light receptor</span>== | ==<span style="color:green;">Green light receptor</span>== | ||
Revision as of 14:20, 18 September 2011
 "
"
 Contact
Contact