Team:Washington/Alkanes/Future/DecarbDesign
From 2011.igem.org
(→Cell Lysis Assay) |
(→Cell Lysis Assay) |
||
| Line 14: | Line 14: | ||
==In Vitro Assay== | ==In Vitro Assay== | ||
| - | === | + | ===Cell Lysis Assay=== |
| - | + | ||
| - | + | ||
===Protein Purification Assay=== | ===Protein Purification Assay=== | ||
Revision as of 19:07, 12 September 2011
Contents |
Background
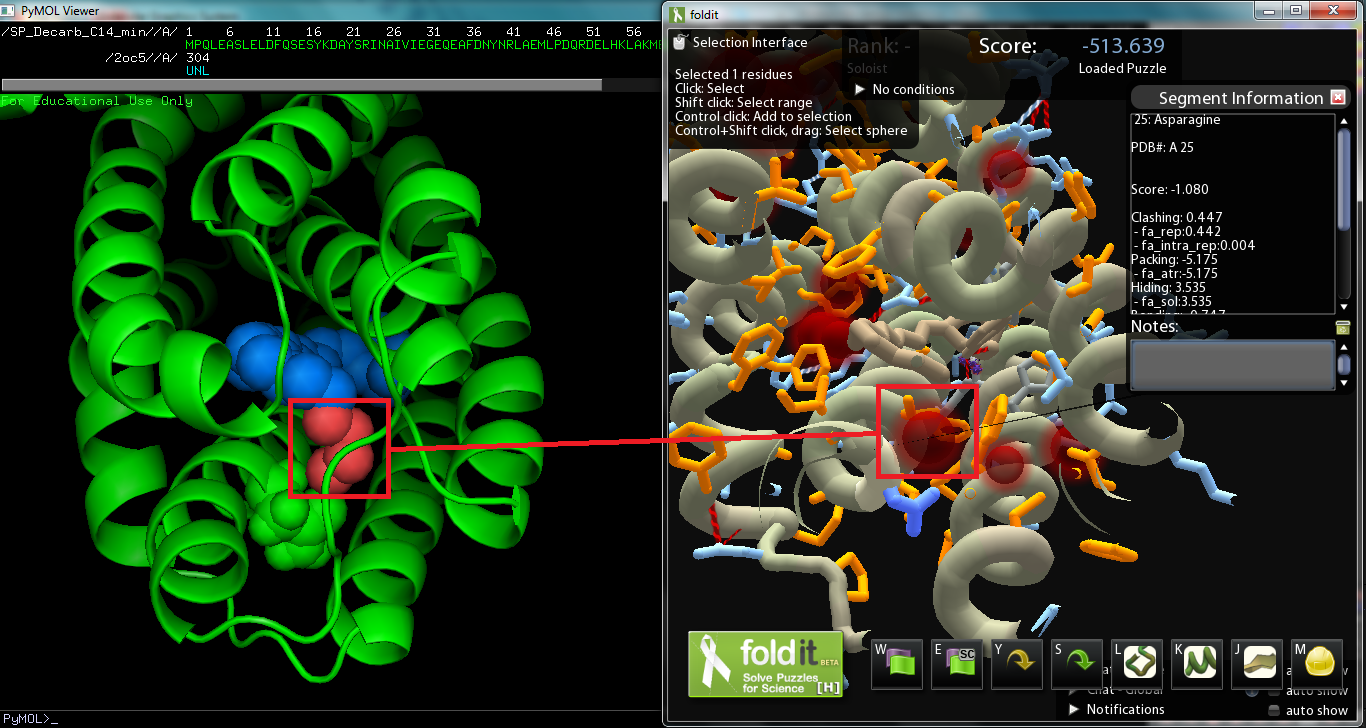
The objective of this subproject was to modify the ADC protein so that we can synthesize a wider variety of alkanes; in other words, mutate the protein so that it works better on a range of saturated fatty aldehydes. The protein was originally surmised to work best on C18 aldehydes, as the crystal structure derived Pymol model of the protein revealed a C18 carboxylic acid bounded to the metal center (though the vector with AAR and ADC proved to work best on C16 aldehydes while less on C14 and C18 aldehydes). Consequently, we decided to strive to modify ADC to work on shorter chain aldehydes, specifically tetradecanal. The substrate on the original Pymol file was modified to model C14 aldehyde, and then the file was converted to a Fold-it puzzle for human interaction. We decided to avoid changing amino acids near the active site, which binds to the aldehyde group, as we wanted to maintain the basic chemistry, decarbonylation (or perhaps deformylation). There was also the issue of the Fold-it modeling 122-Tyrosine hydrogen bonded to the substrate and another side chain, and according to Fold-it's notifications, serine, threonine, and tyrosine cannot "have more than 1 donor and 2 acceptors." As the amino acid we will change will be around the alkyl chain, there are no hydrogen bonds to be made(hydrogen bonds show up on the interface). If we are to make changes so that the protein binds more favorably to C14 aldehydes, we must improve dispersion intermolecular interactions by increasing the surface interaction between protein and substrate. Comparison with the original Pymol file showed that the removal of the four carbon atoms created a spatial “void area.” Targeting the void area with minimal interference or clashes between atoms, the most promising mutation sites are on adjacent sections of two helices positioned opposite of the substrate’s carbon end. The adjacent sections take up amino acids 21 to 25 and 67 to 71, the former of which, being closer, shows promise as a site to mutate to fill in the void area and avoid interferences.
Methods
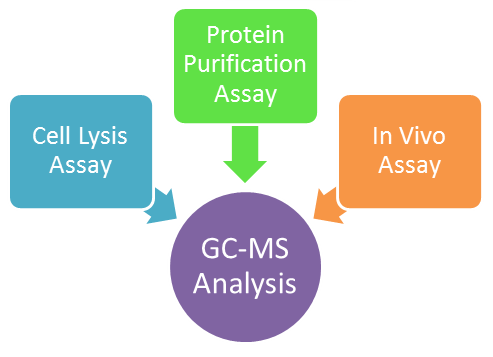
After creating mutations using Fold-It, a computational design software, we developed three different reactions which will be later analyzed using GC-MS to see if our mutated ADC enzyme can produce C13 alkanes. These assays are listed below.
In Vitro Assay
Cell Lysis Assay
Protein Purification Assay
In Vivo Assay
Current Status
- Discuss mutations made and construct submitted
- Show protein gel of expression
- Discuss in vitro assay and how it hasn't worked yet (even for WT)
- Discuss plan for future testing
Parts Submitted
- Plasmids with mutations (ADC_AB4, 1C2, 7A5, 8BE, 9A1, 10B5)
 "
"