Team:Caltech/Week 9
From 2011.igem.org
| (13 intermediate revisions not shown) | |||
| Line 36: | Line 36: | ||
Redid Gibson of pNT002, pNT003, pNT004 with pcrd inserts<br/> | Redid Gibson of pNT002, pNT003, pNT004 with pcrd inserts<br/> | ||
Transformed cells with I732017 and K323089 for some source of lacZ<br/> | Transformed cells with I732017 and K323089 for some source of lacZ<br/> | ||
| + | Re-assembly of DDT gene using PIPE cloning and Gibson, transformed into different cells. | ||
| + | Begin time-dependent biofilm optimization experiment with crystal violet stain. | ||
===Results=== | ===Results=== | ||
I732019 and I732005 plates had no colonies<br/> | I732019 and I732005 plates had no colonies<br/> | ||
pNT004 Gibson plates had no colonies<br/> | pNT004 Gibson plates had no colonies<br/> | ||
CPCR of pNT002 colonies had no insert bands<br/> | CPCR of pNT002 colonies had no insert bands<br/> | ||
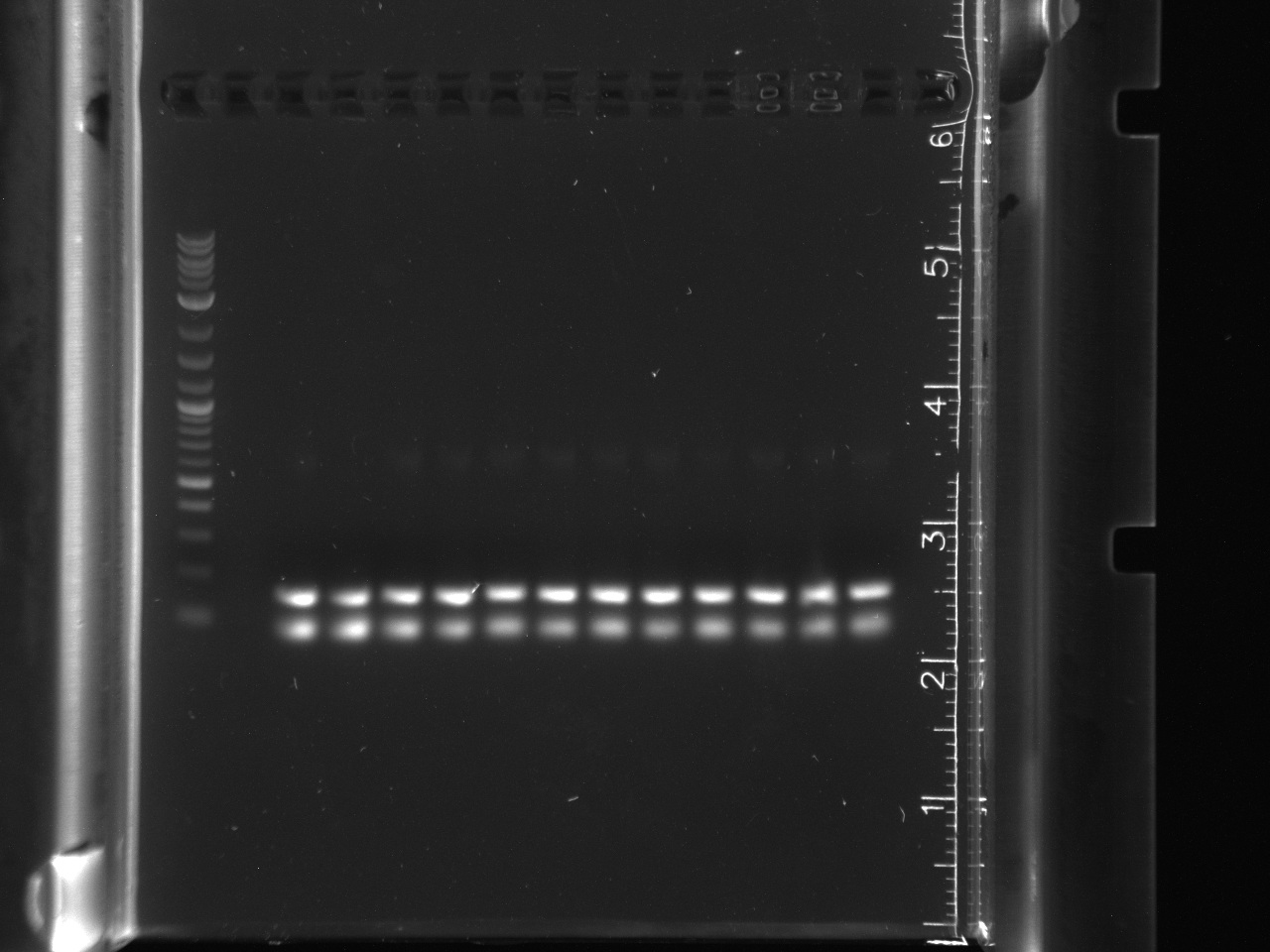
| + | DDT assembly transformations showed colonies<br/> | ||
| + | ==August 11== | ||
| + | Colony PCR of DDT gene colonies<br/> | ||
| + | Transformation of I732005, I732017, I732019 in new XL-10 cells<br/> | ||
| + | PCR of pSB4A5, pSB3K3, pNT002 insert, pNT003 insert. Tried PCA to assemble inserts more specifically.<br/> | ||
| + | Continue time-dependent biofilm optimization experiment with crystal violet stain. | ||
| + | ===Results=== | ||
| + | [[File:DDT_assembly_cpcr.jpg|thumb|left|CPCR of DDT gene]] | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Plate</th> | ||
| + | <th>number of colonies</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 + top ten</td> | ||
| + | <td>3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 - top ten</td> | ||
| + | <td>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT003+ top ten</td> | ||
| + | <td>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT003 - top ten</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT004 + top ten</td> | ||
| + | <td>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT004 - top ten</td> | ||
| + | <td>1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>I372017</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>K323089</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Name</th> | ||
| + | <th>concentration (ng/ul)</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB4A5</td> | ||
| + | <td>57.1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB3k3 1</td> | ||
| + | <td>60.4</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB3k3 2</td> | ||
| + | <td>31.1</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 insert</td> | ||
| + | <td>136.9</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT003 insert</td> | ||
| + | <td>72.0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 insert PCA</td> | ||
| + | <td>15.7</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | ==August 12== | ||
| + | Continue time-dependent biofilm optimization experiment with crystal violet stain.<br/> | ||
| + | PCR of 16s. | ||
| + | ===Results=== | ||
| + | Biobrick transformation plates | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Plate</th> | ||
| + | <th>Colonies</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>I732005</td> | ||
| + | <td>0</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>I732017</td> | ||
| + | <td>~200</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>I732019</td> | ||
| + | <td>65</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | [[File:8-1216sphusion.jpg|thumb|16s amplification, lane 1 NEB 2-log ladder, lane 2 9-3A, lane 3 9-3B, lane 4 9-3C, lane 5 9-9A, lane 6 9-9B, lane 7 9-9C, lane 8 10-3A, lane 9 10-3B, lane 10 10-3C, lane 11 10-6A, lane 12 10-6B, lane 13 10-6C, lane 14 Control 1, lane 15 Control 2]] | ||
| + | ==August 13== | ||
| + | ===Results=== | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th>Plate</th> | ||
| + | <th>Colonies</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 +</td> | ||
| + | <td>~320</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT002 -</td> | ||
| + | <td>~800</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT003+</td> | ||
| + | <td>~500</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pNT003-</td> | ||
| + | <td>~400</td> | ||
| + | </tr> | ||
| + | </table><br/> | ||
| + | |||
| + | Crystal violet biofilm results: | ||
| + | <table border="1"> | ||
| + | <tr> | ||
| + | <th rowspan="2">'''Starting OD'''</th> | ||
| + | <th colspan="2">8/10</th> | ||
| + | <th colspan="3">8/11</th> | ||
| + | <th colspan="3">8/12</th> | ||
| + | <th colspan="2">8/13</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5pm</td> | ||
| + | <td>11pm</td> | ||
| + | <td>11am</td> | ||
| + | <td>5pm</td> | ||
| + | <td>11pm</td> | ||
| + | <td>11am</td> | ||
| + | <td>5pm</td> | ||
| + | <td>11pm</td> | ||
| + | <td>11am</td> | ||
| + | <td>5pm</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>'''0.118'''</td> | ||
| + | <td>.065</td> | ||
| + | <td>.171</td> | ||
| + | <td>.114</td> | ||
| + | <td>.135</td> | ||
| + | <td>.266</td> | ||
| + | <td>.149</td> | ||
| + | <td>.133</td> | ||
| + | <td>.100</td> | ||
| + | <td>.408</td> | ||
| + | <td>.740</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>'''0.170'''</td> | ||
| + | <td>.065</td> | ||
| + | <td>.132</td> | ||
| + | <td>.082</td> | ||
| + | <td>.190</td> | ||
| + | <td>.159</td> | ||
| + | <td>.439</td> | ||
| + | <td>1.875</td> | ||
| + | <td>.106</td> | ||
| + | <td>.323</td> | ||
| + | <td>.545</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>'''0.068'''</td> | ||
| + | <td>.065</td> | ||
| + | <td>.106</td> | ||
| + | <td>.184</td> | ||
| + | <td>.198</td> | ||
| + | <td>.210</td> | ||
| + | <td>.121</td> | ||
| + | <td>.201</td> | ||
| + | <td>.145</td> | ||
| + | <td>.039</td> | ||
| + | <td>.881</td> | ||
| + | </tr> | ||
| + | </table><br/> | ||
| + | <gallery> | ||
| + | File:Biofilm_graphs.jpg|Biofilm experiment chart | ||
| + | File:96-well.jpg|Biofilm experiment 96-well plate | ||
| + | File:Cuvettes.jpg|Biofilm experiment cuvettes | ||
| + | </gallery> | ||
}} | }} | ||
Latest revision as of 18:50, 8 September 2011
|
Project |
August 8Testing for biofilms grown on 96-well plate and Erlenmeyer flasks with crystal violet ResultsBacteria-incubated well remained cystal violet stained after several rinses, indicating the presence of a biofilm, in contrast to the clear control well. Got successful amplification of 16s in LA River sample 9-9. In both gels, the controls had no bands and all other sample lanes had no bands. August 9Evaporate-centrifuge BPA sample to concentrate for GCMS ResultsI732019 and I732005 plates had no colonies. August 10Minimal media transfers ResultsI732019 and I732005 plates had no colonies August 11Colony PCR of DDT gene colonies Results
August 12Continue time-dependent biofilm optimization experiment with crystal violet stain. ResultsBiobrick transformation plates
August 13Results
Crystal violet biofilm results:
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"