Team:BU Wellesley Software/Notebook/KyleNotebook
From 2011.igem.org
| Line 369: | Line 369: | ||
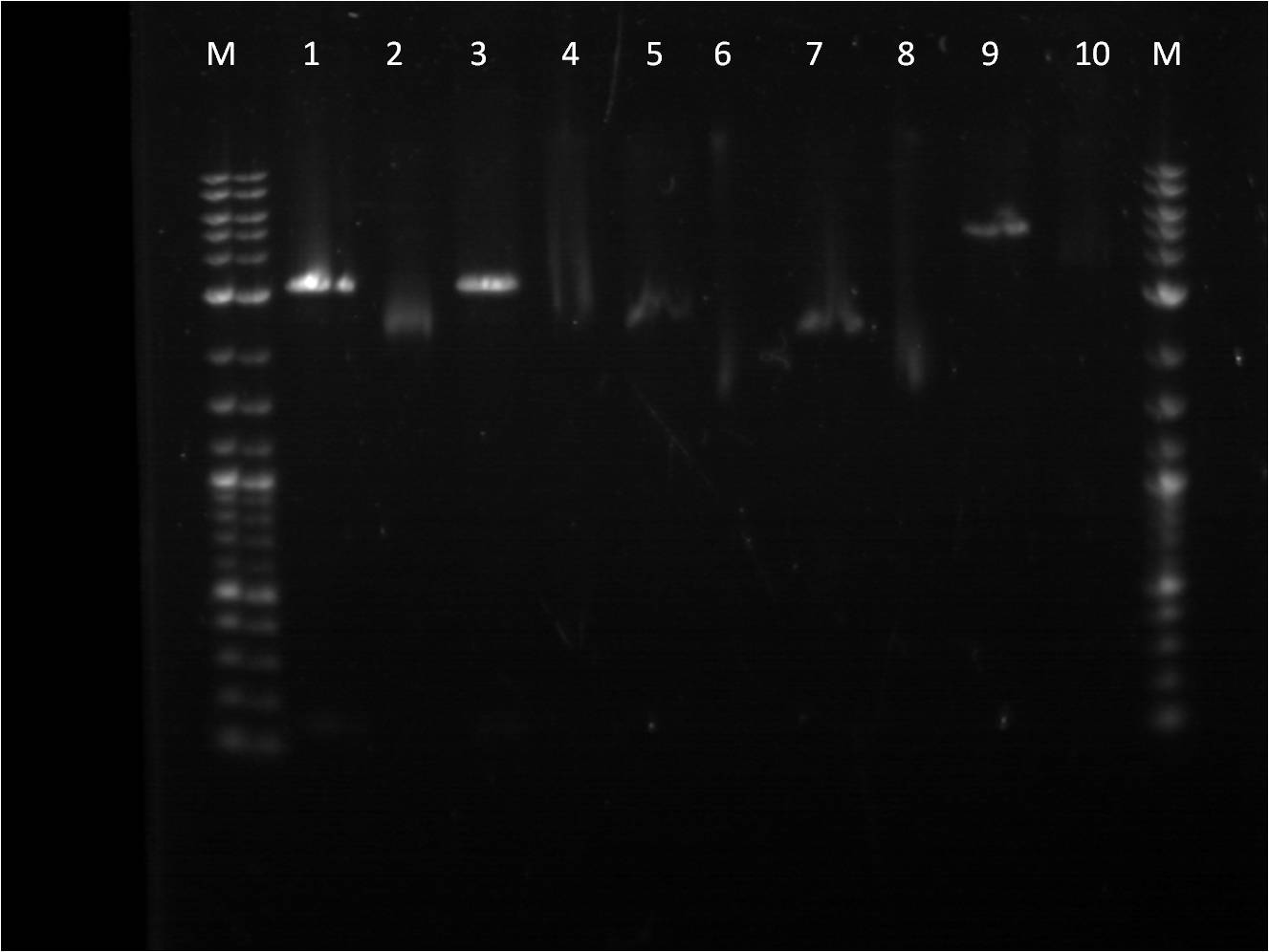
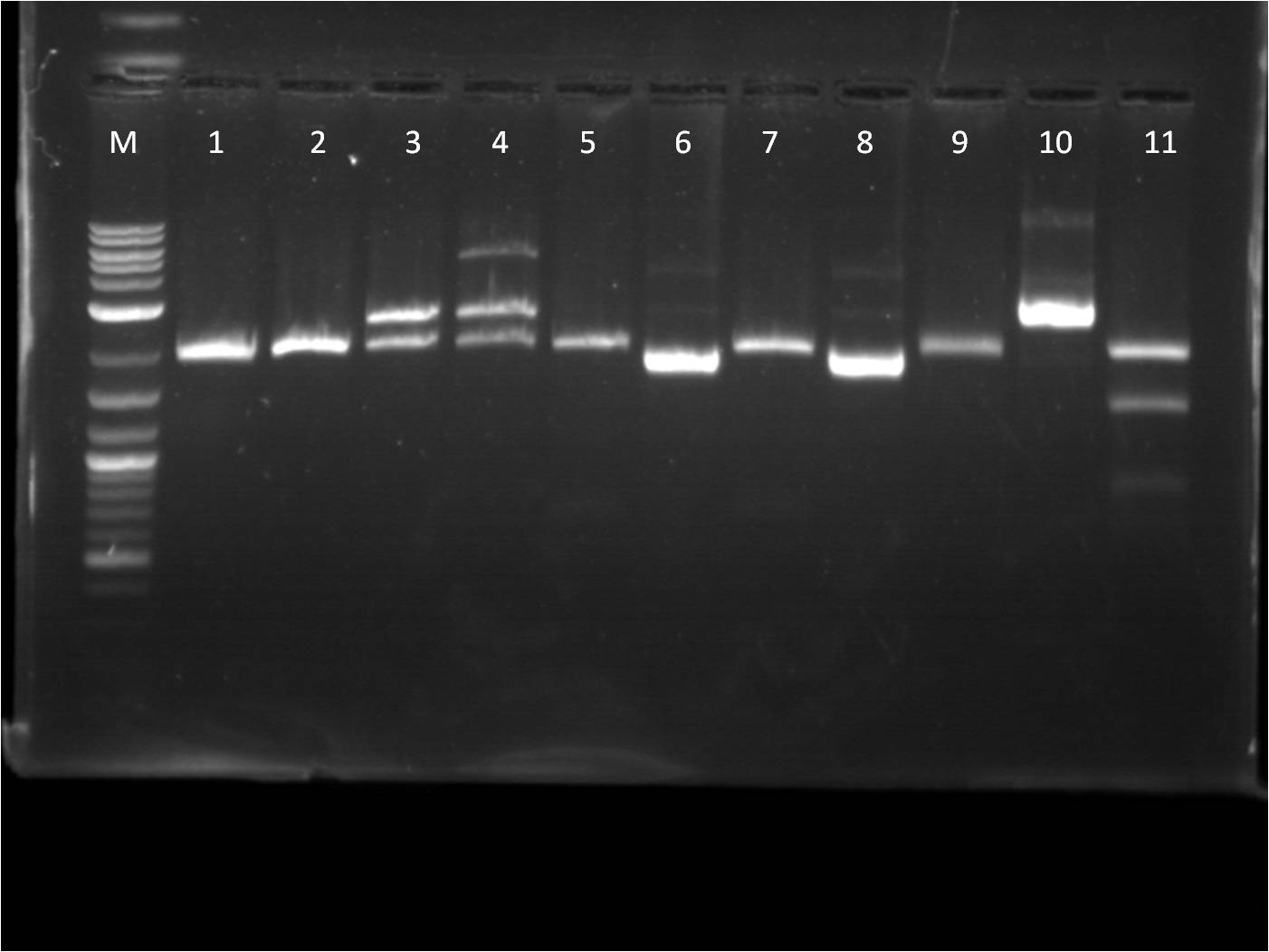
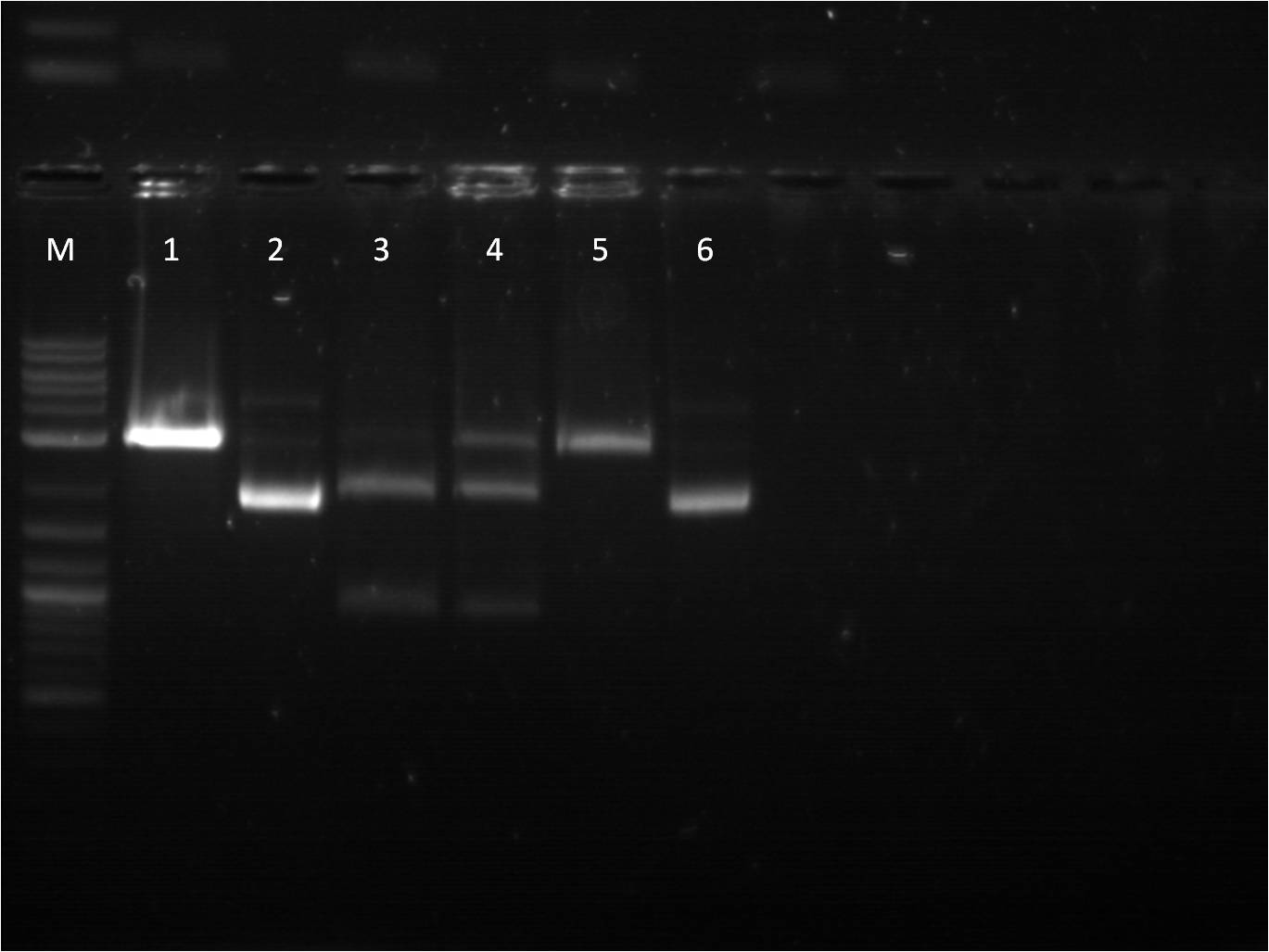
8/17: Last night I set up 4 overnight cultures of BFP+Term in hopes that at least one of them would be the right plasmid. Today, I did a restriction digest of those plasmids, as well as our pTet+G/R/Y_FPc parts in preparation to combine them with my TetR+promoter plasmid. The pTet+_FPc parts were cut with both X,P and E,X so that I do two different ligation reaction with each part: one with the pTet+_FPc as the backbone and one with the pConst+TetR backbone. On the gel, the BFP+Term RD looked exactly like before, having a very small insert(100-200 bp). The pTet+_FPc RD looked great and the DNA was extracted. The rest of the day was spent doing a CIP reaction with the B0015 terminator so that we can avoid having the backbone re-ligate on itself. | 8/17: Last night I set up 4 overnight cultures of BFP+Term in hopes that at least one of them would be the right plasmid. Today, I did a restriction digest of those plasmids, as well as our pTet+G/R/Y_FPc parts in preparation to combine them with my TetR+promoter plasmid. The pTet+_FPc parts were cut with both X,P and E,X so that I do two different ligation reaction with each part: one with the pTet+_FPc as the backbone and one with the pConst+TetR backbone. On the gel, the BFP+Term RD looked exactly like before, having a very small insert(100-200 bp). The pTet+_FPc RD looked great and the DNA was extracted. The rest of the day was spent doing a CIP reaction with the B0015 terminator so that we can avoid having the backbone re-ligate on itself. | ||
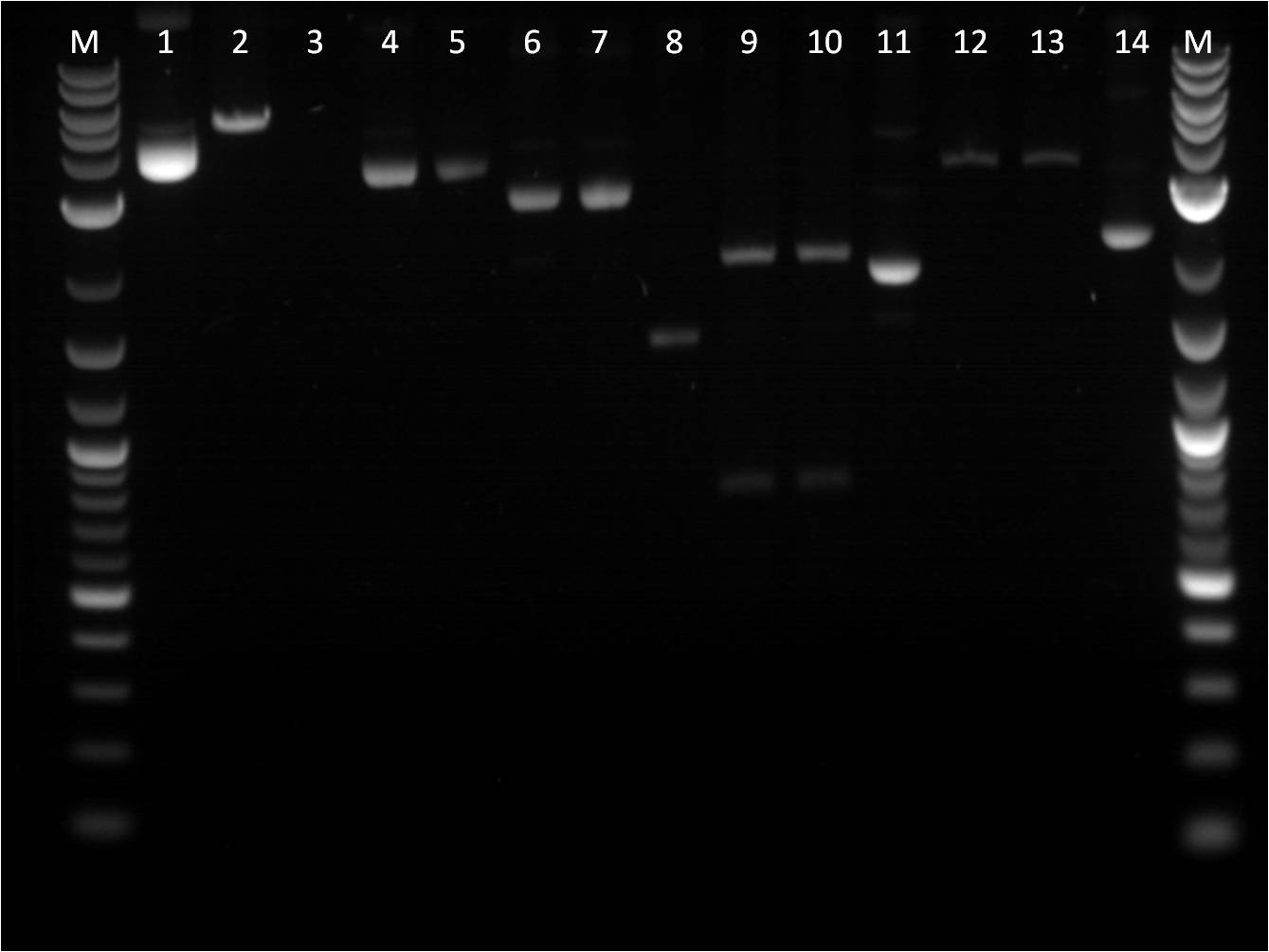
| - | gel | + | [[File: 8_17_11_KJ_RD_top_labeled.jpg | 250px | right|thumb| 8/3/11 Top gel of BFP+Term and pTet+_FPc RD's]] |
| + | |||
| + | TOP | ||
| + | M: Ladder. 1: BFP+Term.1 RD 2: BFP+Term.1 uncut 3: BFP+Term.2 RD 4: BFP+Term.2 uncut | ||
| + | 5: BFP+Term.3 RD 6: BFP+Term.3 uncut 7: BFP+Term.4 RD 8: BFP+Term.4 uncut | ||
| + | 9:pTet+GFPc X,P 10: pTet+GFPc E,X 11: pTet+GFPc uncut | ||
| + | |||
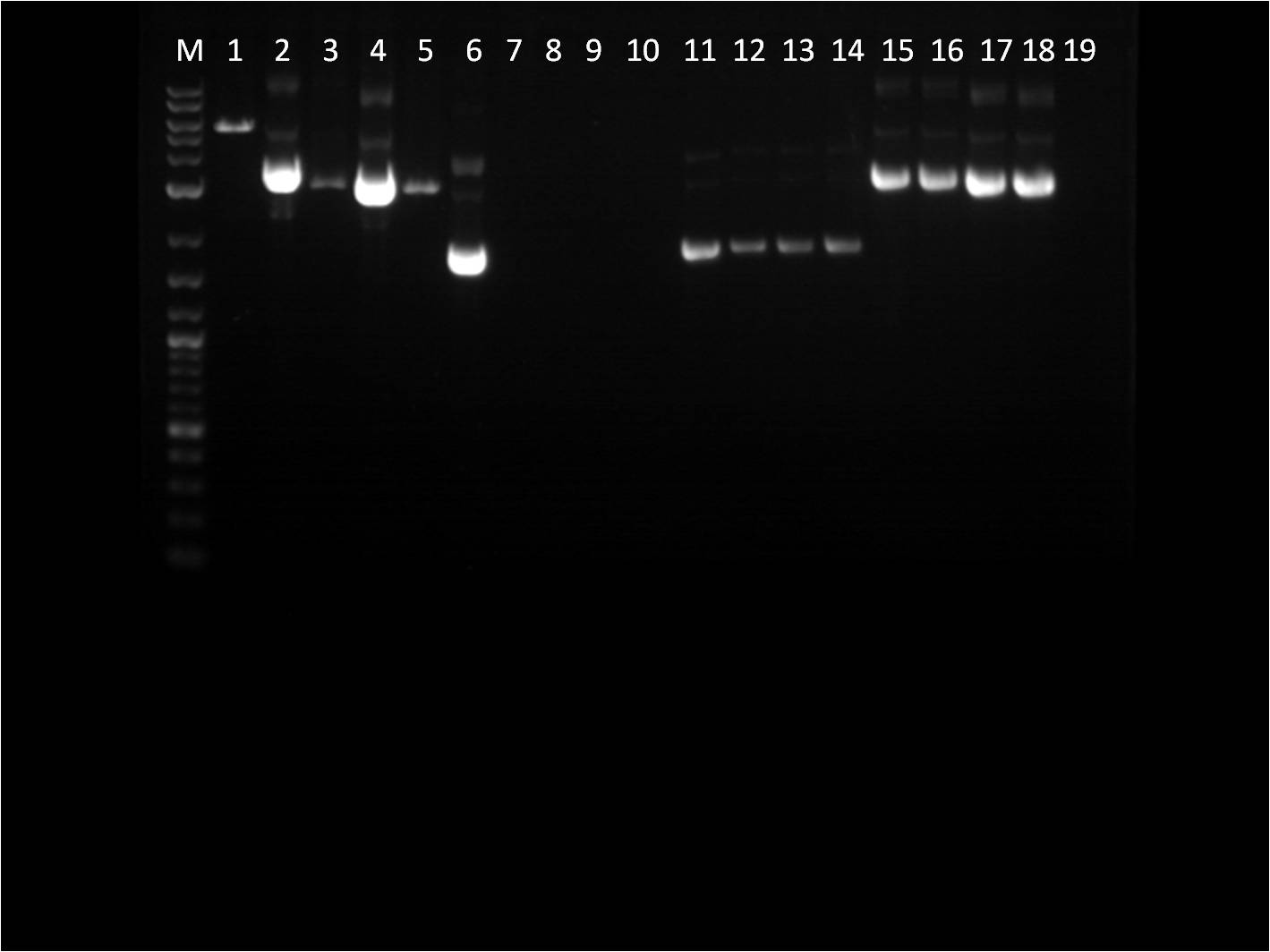
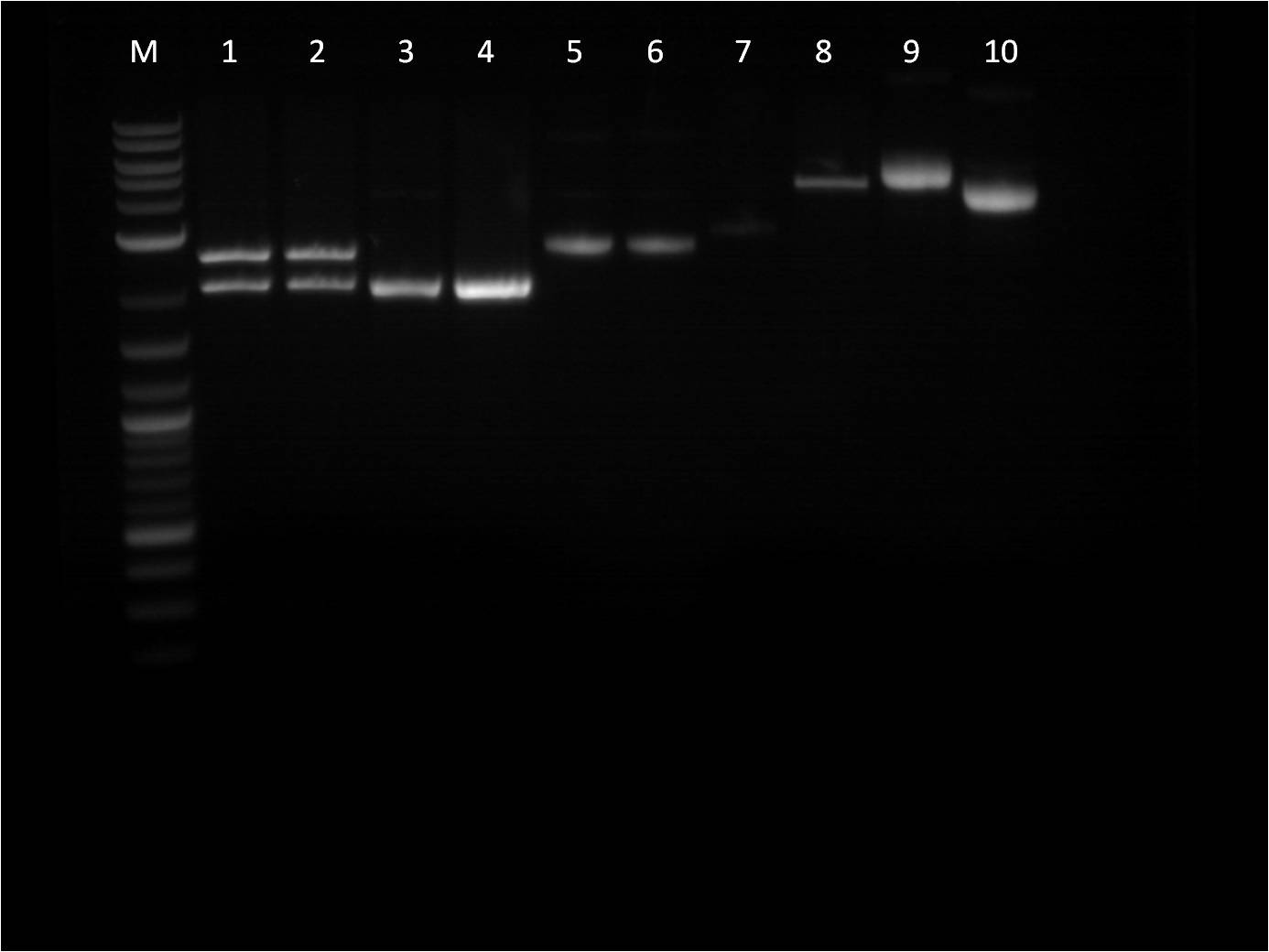
| + | [[File: 8_17_11_KJ_RD_bottom_labeled.jpg | 250px | left|thumb| 8/3/11 Bottom gel of BFP+Term and pTet+_FPc RD's]] | ||
| + | |||
| + | BOTTOM | ||
| + | M: Ladder. 1: pTet+YFPc E,X 2: pTet+YFPc uncut 3: pTet+YFPc X,P 4: pTet+RFPc X,P | ||
| + | 5: pTet+RFPc E,X 6: pTet+RFPc uncut | ||
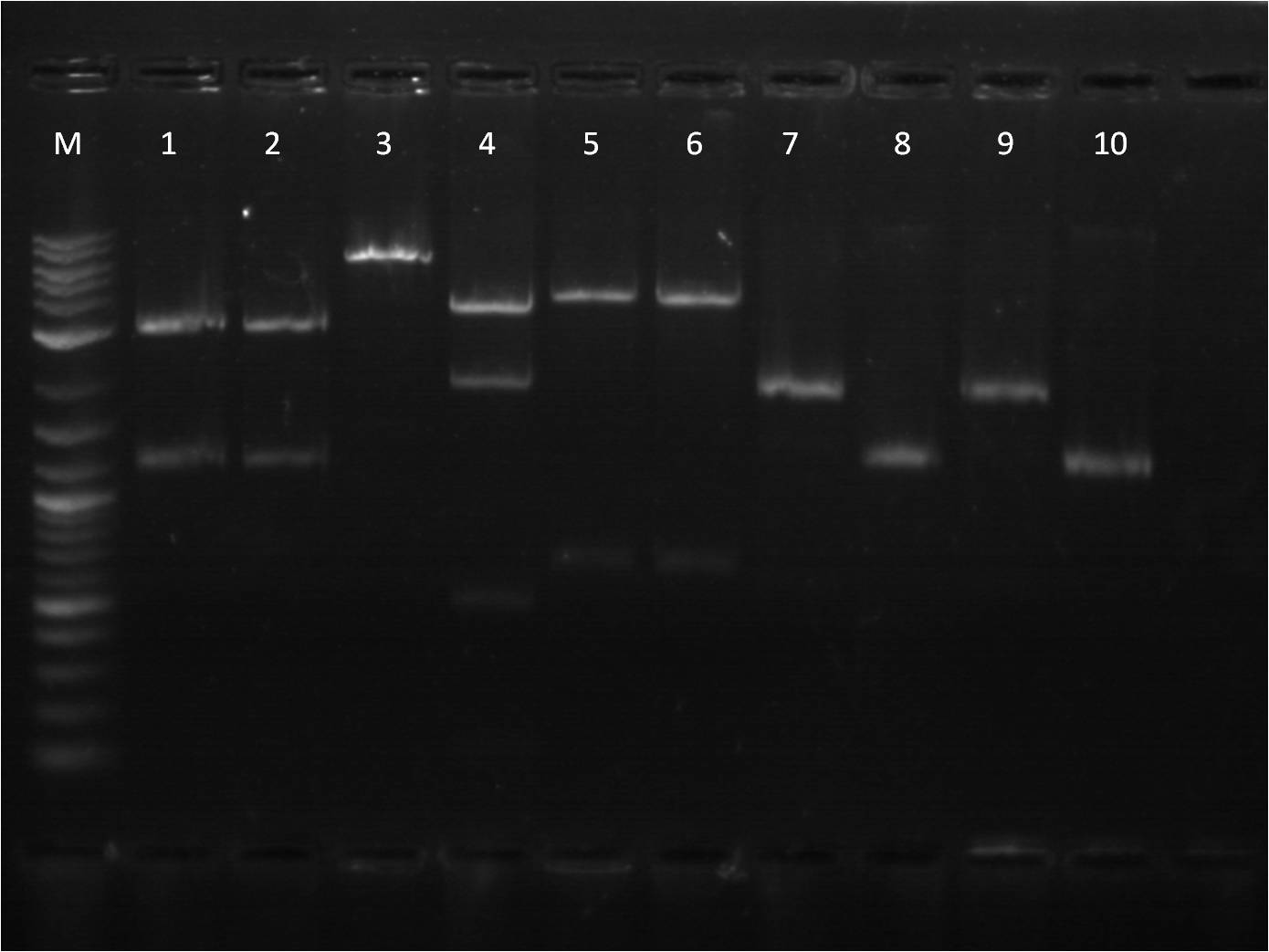
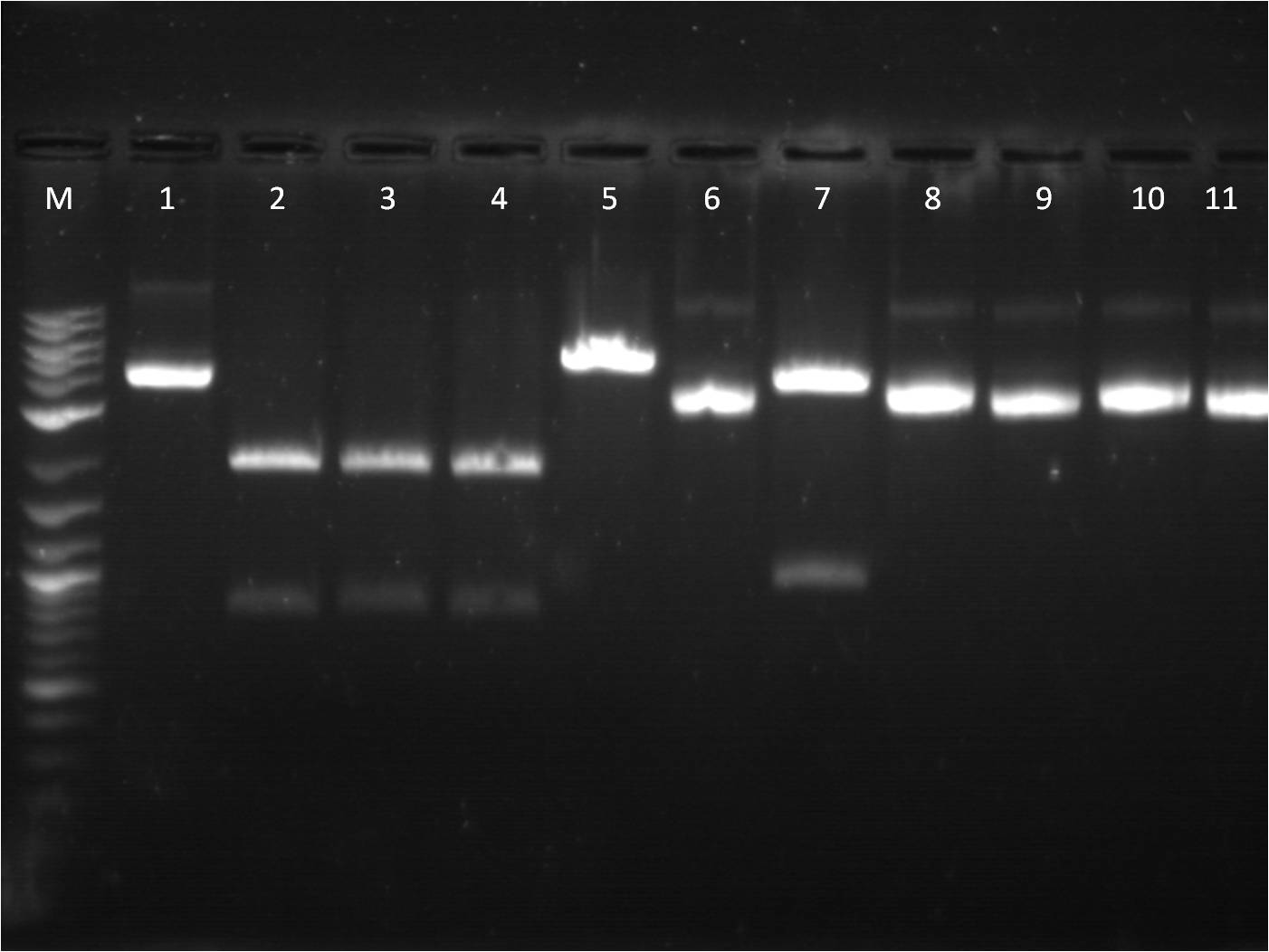
8/18: Today I ligated and transformed BFP and the CIP'd backbone, B0015 (Terminator). Our restock of SpeI enzyme just came today, so I did a restriction digest on my TetR+pCat (P0440+I14033), cutting at S,P and E,S so that I can both use it as an insert and backbone. | 8/18: Today I ligated and transformed BFP and the CIP'd backbone, B0015 (Terminator). Our restock of SpeI enzyme just came today, so I did a restriction digest on my TetR+pCat (P0440+I14033), cutting at S,P and E,S so that I can both use it as an insert and backbone. | ||
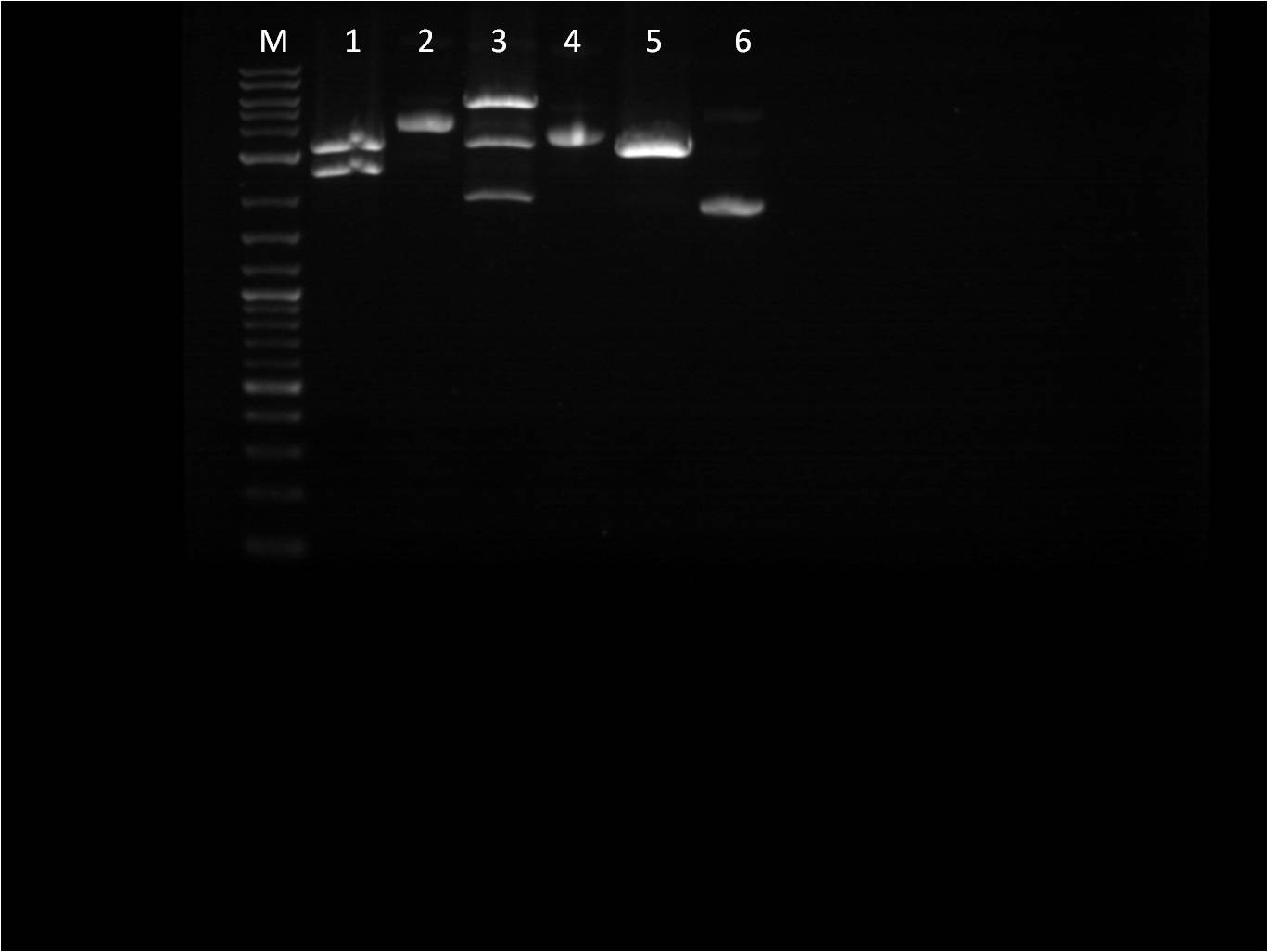
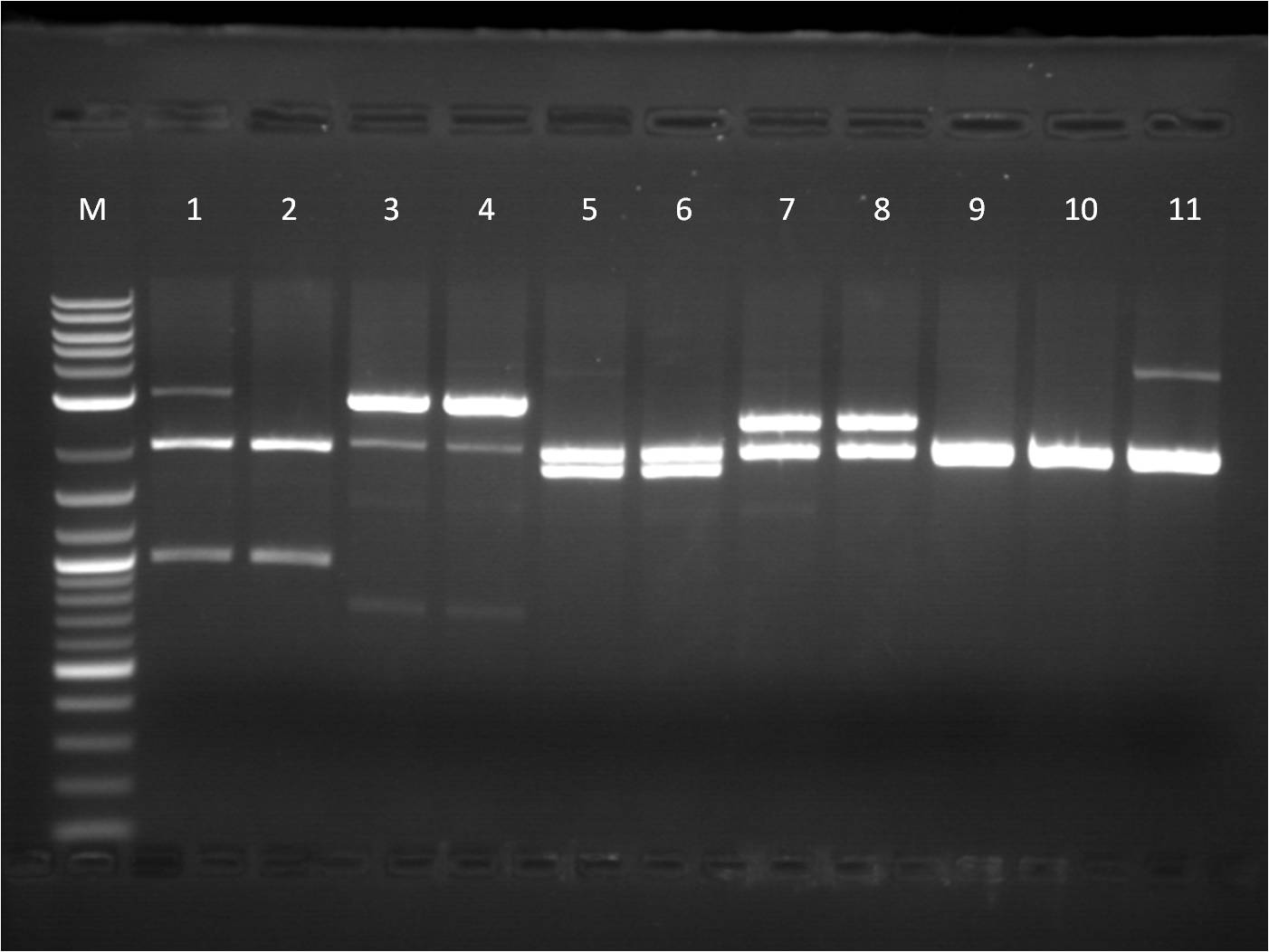
| - | + | [[File: 8_18_11_RD_VYKJ_bottom_labeled.jpg | 250px | right|thumb| 8/3/11 Gel of TetR+pCat]] | |
| + | |||
| + | M: Ladder. 5: pCat.1+TetR.1 S,P 6: pCat.1+TetR.1 uncut 7: pCat+TetR.3 E,S 8: pCat+TetR.3 uncut | ||
| + | 9: pCat.1+TetR.1 uncut 10: pCat.1+TetR.3 uncut 11: pCat.3+TetR.1 uncut | ||
| + | |||
8/19: After extracting my cut TetR+pCat DNA, I ligated it with the pTet+_FPc parts. I made two samples of each, one using the TetR+pCat as the backbone and the other using the pTet+_FPc as the backbone. I also ligated the TetR gene with another constitutive promoter, J23101. Both these ligations were then transformed. The transformations of BFP+Term (CIP) worked well. Today, we also receive a new BFP part, K156026, from the parts registry that we had to request. | 8/19: After extracting my cut TetR+pCat DNA, I ligated it with the pTet+_FPc parts. I made two samples of each, one using the TetR+pCat as the backbone and the other using the pTet+_FPc as the backbone. I also ligated the TetR gene with another constitutive promoter, J23101. Both these ligations were then transformed. The transformations of BFP+Term (CIP) worked well. Today, we also receive a new BFP part, K156026, from the parts registry that we had to request. | ||
Revision as of 13:57, 5 September 2011

Wet Lab Team Notebook - Kyle Jones
WEEK 1: 6/6-6/10/2011 |
|
As the first week of actual iGEM work, there is some initial prep work that needs to be done. Before we can start focusing on the main project involving the development of DNA plasmids and invertases to function within the cell, we must confirm our protocols and create working plasmids with fluorescent protein genes. Also, we want to see which promoters and ribosomal binding sites (RBS) work the best together. 6/6: Monday was our last day of "boot camp". With the comp. team and Wellesley students, we discussed basic synthetic biology and the different procedures/protocols that we would be using in the lab. We also reviewed a paper from Science magazine, Synthetic Gene Networks that Count by Friedland, et al. A tour of the lab concluded the day. 6/7: In the first day in the lab, I did a liquid culture plasmid prep of plated transformations that our Post-Doc mentor Traci had already done. These would go into the incubator and be ready for mini-prep and plasmid isolation tomorrow. As a wet lab team, we browsed the Registry of Standard Biological Parts and chose the parts that we would work with over the next couple of weeks. In all, we chose 3 constitutive promoters, 2 inducible promoters, and 3 RBS, each of varying strength. We subsequently went through the transformation protocol and inserted the plasmids into competent cells. These would incubate overnight. 6/8: Today I did mini-prep and plasmid isolation on the liquid cultures from the day before. The DNA quantification of these samples proved to be poor, with only a few having viable concentrations of DNA for further use. Additionally, we ran a gel electrophoresis of the plasmid samples. The gel showed no bands of DNA, thus further confirming the absence of viable plasmid DNA from the samples. This result is causing some consternation as the exact reason for the problem is still unknown to us. The transformation plates of our selected parts were successful. The rest of the day was spent doing liquid culture plasmid preps of these plates that we had transformed on 6/7. 6/9: Today we did the mini-prep and plasmid isolation of our transformation of selected parts(3 const. promoters, 2 ind. promoters, and 3 RBS). The DNA quantification of these samples were poor (like our samples the day before) and a gel electrophoresis of the plasmids gave indication of little to no plasmid DNA. Only one part (UV light promoter) had a viable DNA concentration. One thing we noticed is that the samples did not grow well in the liquid cultures and there was almost a stringy characteristic to the cells in the tube. For further investigate, we went to the Biology labs and performed Gramm Staining on 6 of the samples. What we saw under the microscope was some cells that resembled E. Coli along with bacteria in a long string/beadlike form. Our conclusion was that the samples were contaminated and that we needed to be more conscientious about lab cleanliness. 6/10: There was not much work in the lab today. I autoclaved some pipet tips that we would use. Instead, the day was spent organizing how we would put together our different BioBrick parts into a full working construct plasmid. This included how to take out the promoters in the original RBS plasmids, as well as which restriction enzymes to use. We planned out the next week and assigned tasks to everyone. |
WEEK 2: 6/12-6/16/2011 |
|
This week, as a team, we will combine the GFP gene and terminator into one plasmid. Also we will continue to troubleshoot and figure out what went wrong with our transformations from Week 1. If we need to, we will set up new transformations of our selected BioBrick parts.
6/14: The transformations from yesterday grew overnight and I checked them this morning. The negative control plate we did (T10 E. coli cells only) grew a lot of colonies, thus effectively pinpointing the source of our problems the past week. The agar plates which had ampicillin on them were no longer effective in killing all other bacteria besides what we had transformed which carried resistance to the antibiotic. Hence why we had such contamination and poor isolation of plasmid DNA. The other plates looks abnormal for E. coli colonies and they were thrown out. The Kanamycin resistant plates worked fine and had no contamination. We proceeded with plasmid preps for the two samples that were grown on Kanamycin plates, the GFP+Terminator (BBa_J52028+ BBa_B0015) composite part and the constitutive promoter BBa_I14033.
6/16: This morning the transformation plates of the 5 promoters, 3 RBS, and RFP looked good. Most had a few colonies and some had a lot. Only the inducible UV light promoter (BBa_I765001) had no colonies (as well as the negative controls). Liquid culture plasmid preps were done on the plates that grew. In attempt to make more GFP+Term composite parts which we will need to combine with the various RBS and promoter parts, I did a restriction digest, gel electrophoresis, and gel extraction. It will be ligated later today and then transformed along with our new parts tomorrow. |
WEEK 3: 6/20-6/24/2011 |
|
6/20: This morning I set up the QIAcube to do minipreps of the liquid plasmid cultures of our transformed reporters (RFP BBa_E1010, EYFP BBa_E0032, ECFP BBa_0020, BFP BBa_K156010) and composite parts (GFPc BBa_E0240 and YFPc BBa_E0430) as well as a promoter (BBa_R2000) chosen for the composite parts. The DNA quantification was good, with most numbers around 30-50 ng/ul. In anticipation of needing more promoter and RBS plasmid DNA for our constructs, I used the transformation plates from 6/15/11 to make liquid plasmid preps. The rest of the afternoon was spent planning for our BU iGEM meeting and what we would present as a wet lab team.
6/21: This morning I prepared minipreps of the promoter and RBS liquid cultures from yesterday for the QIAcube to run. Today was our BU iGEM wet and dry lab meeting. As our first one, it involved an overall presentation on what has been happening in each of the teams. As a wet lab team, our presentation was more group as a lot of what we had done, everyone had been apart of. We discussed our initial contamination problems, the parts that we have chosen, and our current success. Also we developed a timeline of events: June -4 “devices” PCR TB Genes (made-tested-verif/Clotho) -Teams (assigned, robot learning, RD/ligation) -Picked “n” invertase (have) -PCR TB Genes July -Get invertase ready -Make 4 devices with invertase... Test -Robot puts together devices (RD/ligation 2 parts) -Manual Reconfig -Robot puts devices together -Robot Transformation 6/22: Today I set up a gel electrophoresis of restriction digest samples that the rest of the wet lab team had set up the day before (see notebooks of Alberto, Margeux, and Vanessa). The samples were E0430 (YFP composite), E0240 (GFP composite), R2000 (constitutive promoter), and B0015 (terminator). Each sample took two wells and were followed by their uncut plasmids.
6/23: This morning I did restriction digest of our GFP+term composite part, J61101 (RBS), J61100 (RBS), J61127 (RBS), and K156010 (BFP). The RBS's I cut with Spe1 and Pst1 while the BFP I cut with EcoR1 and Spe1. The GFP+term was cut with Xba1 and Pst1. A gel electrophoresis was run. The GFP+term still only showed one band, the same result we had two days earlier. Thus we decided that the restriction digest was not the conflict here and rather the ligation did not work properly. With the BFP and RBS samples, the separation looked good and the samples ( lower bands for BFP corresponding to the smaller insert and higher bands for RBS corresponding to the large backbone) were cut out and purified through gel extraction quickprep on the QIAcube.
M: Ladder. 1/2: E1010. RD 3: E1010 uncut. 4/5: E0032 RD. 6: E0032 uncut. 7/8: E0020 RD. 9: E0020 uncut. 10/11: K156010 RD. 12: K156010 uncut. 13/14: J61101 RD. 15: J61101 uncut. 16/17: J61100 RD. 18: J61100 uncut. For some of the uncut plasmids, not enough sample was put into the well, so they are hard to see. For all the fluorescent proteins, the enzymes EcoRI and SpeI were used and thus the lower bands on the gel were removed. The RBS's were cut with SpeI and PstI and the higher bands were removed. Because we were running out of RBS plasmids and the promoter I14033, I used the frozen glycerol stock of our transformed cells to plate and grow overnight for plasmid preps over the weekend. |
WEEK 4: 6/27-7/1/2011 |
|
After a week of plasmid preps and restriction digest, we had the stock of DNA to start ligated and putting together our composite parts or "devices". I am on the "bottoms up" team in that we will be building devices from the bottom up, starting with combining the fluorescent protein and terminator, then add an RBS and promoter. 6/27: Some members of the wet lab were having problems with ligation, so we looked at our formula for calculating the amount of insert and backbone DNA needed. The ratio of insert to backbone should be about 2-6 to 1 and ratio of base pairs has to be factored in such that Nanogram (ng)of inserts = (ng of vector x size of insert)/size of the vector x (insert : vector molar ratio) Using this formula and a 6:1 ratio of insert to backbone, I performed a ligation of RFP, ECFP, EYFP, and BFP with terminator. Instead of running the reaction for an hour at 16 C, I had the ligation run at room temperature for 30 minutes and then heat inactivated the reaction at 65 C for 10 minutes. Following the ligation protocol, I did a transformation of the ligated mixture into competent cells. 6/28: The plates from yesterday's ligation/transformation did not grow anything. This is increasing our troubles with our ligation protocol. Something is either going wrong with our ligation or restriction digest of the plasmids. However, since we are still seeing separation and appropriate band size in gel electrophoresis a from restriction digest, I think that it is most likely a problem with our ligation procedure. So today I did a ligation of RFP and BFP with terminator. This time I set up two reactions for each, one to run at 16 C for 1 hour and one at 4 C overnight. The hope was that an increased reaction time for the ligation would slow down the reaction and provide more time and a better ligation. An increased amount of ligase enzyme ( 2 ul instead of 1 ul) was also used. 6/29: Today I took the ligation reactions from yesterday (RFP + Term and BFP + Term) and did transformations with them. After I was half way through the transformation protocol, I realized that I had not heat inactivated the ligation mixtures from the 4 C overnight. Thus, I took what was left in those tubes, heat inactivated at 65 C for 10 mins, and then transformed them into competent cells. The volume of ligation reaction was 4.5 ul and 7.5 ul for RFP+term and BFP+term respectively. This is much less that our usual volume of 10 ul per transformation reaction. i also plated a negative control of competent cells on KAN plate. Today I also took record of our frozen glycerol stock of transformed plasmids in the -20 C freezer. In attempt to organize and optimize lab space, we are setting up boxes and online databases to keep a record of samples we have, their location in these plasmid boxes, and other important information such as concentration and volume. Eventually we would like to have a Clotho tool similar to the current app Batterboard to keep this online database. Right now we are just using Microsoft Excel and google documents, but today we spoke to members of the computational team about modifying Batterboard to include certain features like a 81-box interface rather than the 96-well plate and a search option.
Since our ligation reactions were still no successful, I continued to troubleshoot, looking at the New England Biolabs (NEB), manufacturer of the ligase enzyme, website about certain problems that may arise for ligation procedures: http://www.neb.com/nebecomm/products/productM0202.asp There they suggested using 5ul of ligation reaction per 50 ul of competent cells for transformation. We had been using 10 ul of ligation reaction for our transformation protocols. To test this, I set up a transformation using the ligation reaction of RFP+term from 6/27 and made 3 reactions: 1ul of ligation, 2ul of ligation, and 4ul of ligation reaction. This time I also plated controls of competent cells on KAN plate as well as just LB agar plate. The LB agar plate is meant to show if there is anything wrong with the competent cells, as they should grow well in plain LB. Another thing that we have notice with our ligation protocol is that our calculations are slightly off. Because the volumes and masses of backbone and insert are relative to each other, you must standardize one measurement. Initially we standardized the volume of the backbone, but this resulted in a very low mass of DNA for the ligation reaction. Thus, we changed our calculations so that we standardized the mass of the backbone to 50 ng. This may have been our problem with our ligation protocols. 7/1: The transformations using varying volumes of ligation reaction in the transformation protocol were unsuccessful and grew nothing. This is not a complete surprise after our discovery of wrongful calculations leading to a low mass of DNA in the ligation reactions. However other wet lab members' ligation reactions using the correct calculations of insert and backbone did not grow as well. Thus, we are forced to brainstorm more on what is going wrong. On the NEB website for the protocol using T4 DNA ligase with cohesive (sticky) ends, it suggest a ligation period of 16 C overnight. Thus, I set up two samples of BFP + terminator: one with a 3:1 ratio of insert to vector and the other with a 6:1 ratio. The tubes would sit overnight in the PCR machine and hopefully give us a proper ligation. |
WEEK 5: 7/3-7/7/2011 |
|
7/3: Today I came to just transform the overnight ligation reactions from 7/1. We used only 40 ul of competent cells due to our low stock of cells. Also, 4ul of ligation mixture was used in the transformation protocol. The plates will be left on the bench top for 2 days. 7/4: Holiday! 7/5: This morning I checked on the transformations from 7/3. Every plate had a thin layer of bacteria on them, almost a lawn of bacteria. This is not usual for transformations and I am somewhat skeptical about the success of these plates. Also we have realized that our stock of LB broth is contaminated with mold, therefore we are going to redo these transformations and include a negative control. Today, I also did a restriction digest of my RFP+terminator that I had ligated and transformed last week as well as some promoters that Vanessa needed. The RFP+Term was cut with XbaI and PstI. The nanograms of DNA for the restriction digest was increased from 500ng to 750ng in hopes of getting greater concentrations for our gel extractions. M: ladder. 1: RFP+Term.16 RD 2: RFP+Term.16 uncut. 3: RFP+Term.4 RD. 4: RFP+Term.4 uncut 5: I13453 RD. 6: I13453 uncut. 7: R0040 RD. 8: R0040 uncut 9: I14033 RD. 10: I14033 uncut The gel did not come out well and is very smudgy. This can be from improper placement of the comb that molds the wells when the gel is made as well as too little buffer. Also, the RFP+Term only has one band that is slightly higher than its uncut form. This band is also around 3kbp which indicates that it is not the ligated part that we had hoped for as the part should be around 4-5kbp in length if the backbone had been cut. 7/6: This morning I checked the re-transformed plates from 7/3 using the same samples and same plates. The negative control (T10 cells only on KAN plates) grew culture and all of the plates looked the same. It seems that the plates simply did not contain the antibiotic as all of the growth of bacteria was on the surface. Thus we have to make new KAN plates. In the meantime, I used LB plates and spread a 1% Kanamycin solution on them before using them for my transformations. I used the same samples from 7/3, however I just used the 6:1 ratio samples. 7/7: Our ligation/transformation troubles continued as none of our reactions from 7/6 grew anything. The negative control looked fine and the cells seemed good (still growing on just plain LB). Thus we are almost certain this is a ligation problem and are going to look into buying a new ligation mixture, a strong and quicker one, that will hopefully solve these problems. While we are waiting for the new ligation enzymes, we are going to stock up on plasmids that we have used up or will need. I made plasmid preps with RFP, GFP, and Terminator. Today was also our Joint BU/Wellesley iGEM meeting. Each team (BU Comp, BU Wet, and Wellesley Comp) gave updates on their work so far. We also discussed further collaboration between the teams as well as possible community outreach opportunities. 7/8: Today there is not a whole lot to do. I started off doing mini preps of the plasmid preps from yesterday. Because we had 24 samples, we did half in the QIAcube and half by hand. Also, I wanted to see if there was any difference in the concentration of the plasmid samples from the QIAcube or by hand, so I made two tubes from the same sample. Some values had a significant difference (20 ng/ul difference) while others were pretty close (0.1 ng/ul difference). Therefore, no real conclusion can be made on this matter until more samples are compared. Later this afternoon, the new ligase enzyme, Quick Ligase from NEB arrived. I did one sample of BFP+terminator using the Quick Ligase and then transformed it. The plates will sit out over the weekend and will be checked on Monday. |
WEEK 6: 7/11-7/15/2011 |
|
7/11: The transformation plates using the Quick Ligase mixtures did not grow anything. Thus Traci, Vanessa, and I took a trip over to MIT to take with a Post-Doc of our collaborators in the Weiss Lab about our ligation problems. While their we discussed the possibility that our problems may lie in transformation of the ligation reactions, rather than the ligation reactions themselves. Since we are using our own homemade competent cells, their ability to take up ligated DNA vector, which is not super-coiled, is significantly decreased. Therefore, we borrowed some of the commercial cells that the MIT lab uses. With these new competent cells, we did transformations of our ligation products. We used a slightly different transformation protocol, recovering the heat shocked cells in 1 ml of LB broth in the 37 C, 300rpm shaker. I noticed that the concentration of cells in the MIT batch was much greater than our tubes of cells. In addition to the competency of our cells, this lack of cell concentration in our sample tubes could also be contributing to our unsuccessful transformations. 7/12: The transformations using our cells did not grow, however the transformations with the commercial cells from MIT saw some growth. The two samples that grew were BFP+term and YFPc+R2000 (constitutive promoter). As noted yesterday, the concentration of our cells per sample tube was much lower than the commercial cells when pelleted. So today we concentrated our cells, using beginning volumes of 800 ul, 1200 ul, 1600 ul, and 2000 ul and they were concentrated into 200 ul volume. We used ligation reactions that we had in the freezer. Also I ran one of my gel extractions that had a very low concentration (1.8 ng/ ul) on a gel to see if anything would show up, as another possible reason for unsuccessful ligation is the quality and amount of DNA from the gel extractions. The gel showed nothing, so there was nothing in the sample. It is something that we have to be aware of when the concentration is so low, that there may actually not be any viable DNA in the sample. 7/13: The transformations using the concentrated cells (our own competent cells) did not work. Likewise, only one plasmid prep from 7/12, YFPc+R200, grew. The DNA quantification for this was very low as well. It is looking increasingly obvious that we will need to buy competent cells. We ordered Bioline Gold Efficiency Alpha-Select cells ( http://www.bioline.com/h_prod_detail.asp?itemid=234). Just to see if we were on the cusp of concentrating our cells, I did another transformation using 4000ul of our cells re-suspended in 200 ul of LB. For these, I did YFPc+ pCat, YFPc+pBad, YFPc+pTet, and a positive control of J23101. 7/14: Transformations of the 4000ul concentrated cells did not work, with the exception on one plate that used the 3000ul concentrated cells, GFPc+R2000, grew a few colonies. As we are waiting for the commercial competent cells to come in, I prepared a new ligation of RFP+term and GFP+term, using our standard ligation protocol (30 mins benchtop, no heat inactivation). I also made some plasmid preps of the BFP+term and YFPc+R2000 that had grown using the MIT cells. There are a lot of false positives on the plate and thus I am trying to be more selective with the colonies. 7/15: This morning I did minipreps of the plasmid preps that grew overnight of BFP+term and YFPc+Term. One of the BFP+term liquid cultures grew well while the others were less cloudy than usual for a liquid culture. |
WEEK 7: 7/18-7/22/2011 |
|
7/18-7/20: Out of Boston 7/21: The transformations of our ligation reactions are now working and all of our parts are now ready. I have RFP+term, GFP+term, and BFP+term on plates now. However, I am still going to do a restriction digest, cutting at X and P, of my old BFP+term to see if it works. 7/22: I prepared minipreps of our transformations as well as make glycerol stocks of YFPc + pBad and YFPc+pCat. I did a restriction digest, again cutting at X and P, of GFP+term, RFP+term, and BFP+term so that I can combine it with an RBS/promoter. |
WEEK 8: 7/25-7/29/2011 |
|
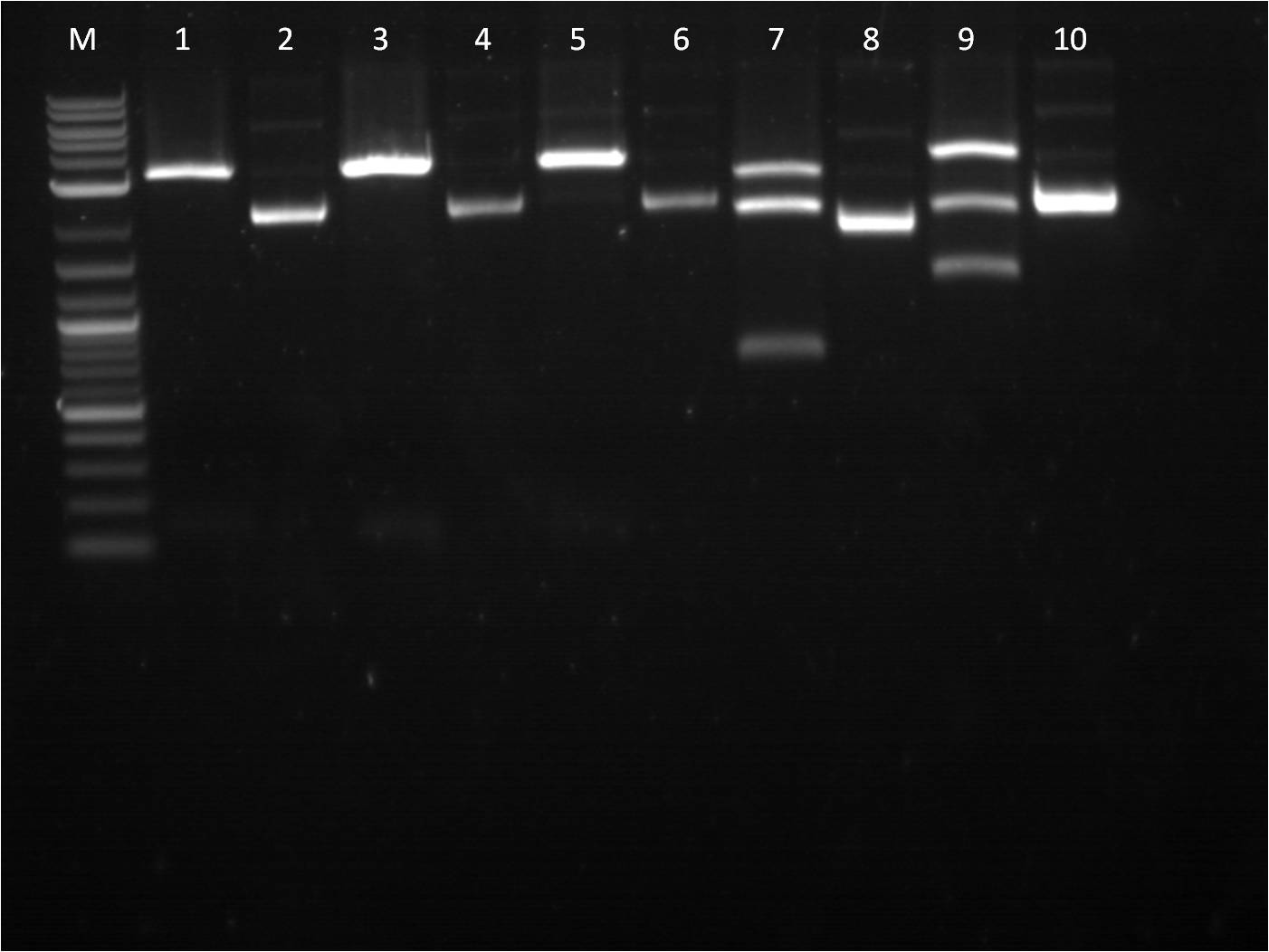
7/25: This morning I ran a gel of my restriction digests from 7/21 and 7/22 of the fluorescent protein genes combined with terminator. M: Ladder. 1: GFP+Term RD 2: GFP+Term uncut 3: RFP+Term RD 4: RFP+Term uncut 5: BFP+Term RD 6: BFP+Term uncut 7: BFP+Term.1 RD 8: BFP+Term.1 uncut 9: BFP+Term.2 RD 10: BFP+Term.2 uncut The inserts from the restriction digest should have been around 1350bp, 810bp, and 849 bp for GFP+Term, RFP+Term and BFP+term respectively. However, on the gel, only one band shows up with the incorrect size. Also, the samples of BFP+term from 7/15 did not show up at all on the gel, thus are not viable samples. With the other samples that only produced one band, the restriction digest must have only cut at one place and did not fully complete. 7/26: Since the restriction digest from 7/25 did not work well, I decided to increase the nanograms of DNA in my reaction from 500ng to 2000ng so that there is more DNA that can possibly be digested. The RD was still run for 2 hours and involved the GFP+term, BFP+term, RFP+term. M: Ladder. 1: GFP+Term RD 2: GFP+Term uncut 3: RFP+Term RD 4: RFP+Term uncut 5: BFP+Term RD 6: BFP+Term uncut Again, the plasmids did not correctly cut. The GFP+term has two bands but of the wrong size while RFP+term has three bands, all of incorrect size. I looked on the NEB website to troubleshoot my restriction digest problems. I found that the double digestion of XbaI and PstI with Buffer 3 does not give 100% efficiency, thus a longer incubation time is necessary to have a better reaction. 7/27: To further troubleshoot my restriction digest problems, I ran the uncut plasmids of GFP+Term, RFP+Term, and BFP+Term against uncut Terminator to make sure there is a difference in length on a gel. It did turn out to look correct in terms of size compared to the DNA ladder and uncut terminator. Thus, I did another restriction digest, using 2000ng of DNA and an incubation period of 3 and 4 hours. 7/28: This morning I ran a gel of the RD's from 7/27 of the _FP's+term.
7/29: The transformations from 7/28 worked. Today is our iGEM camping trip, so the overnight cultures will have to be set up on Monday. |
WEEK 9: 8/1-8/5/2011 |
|
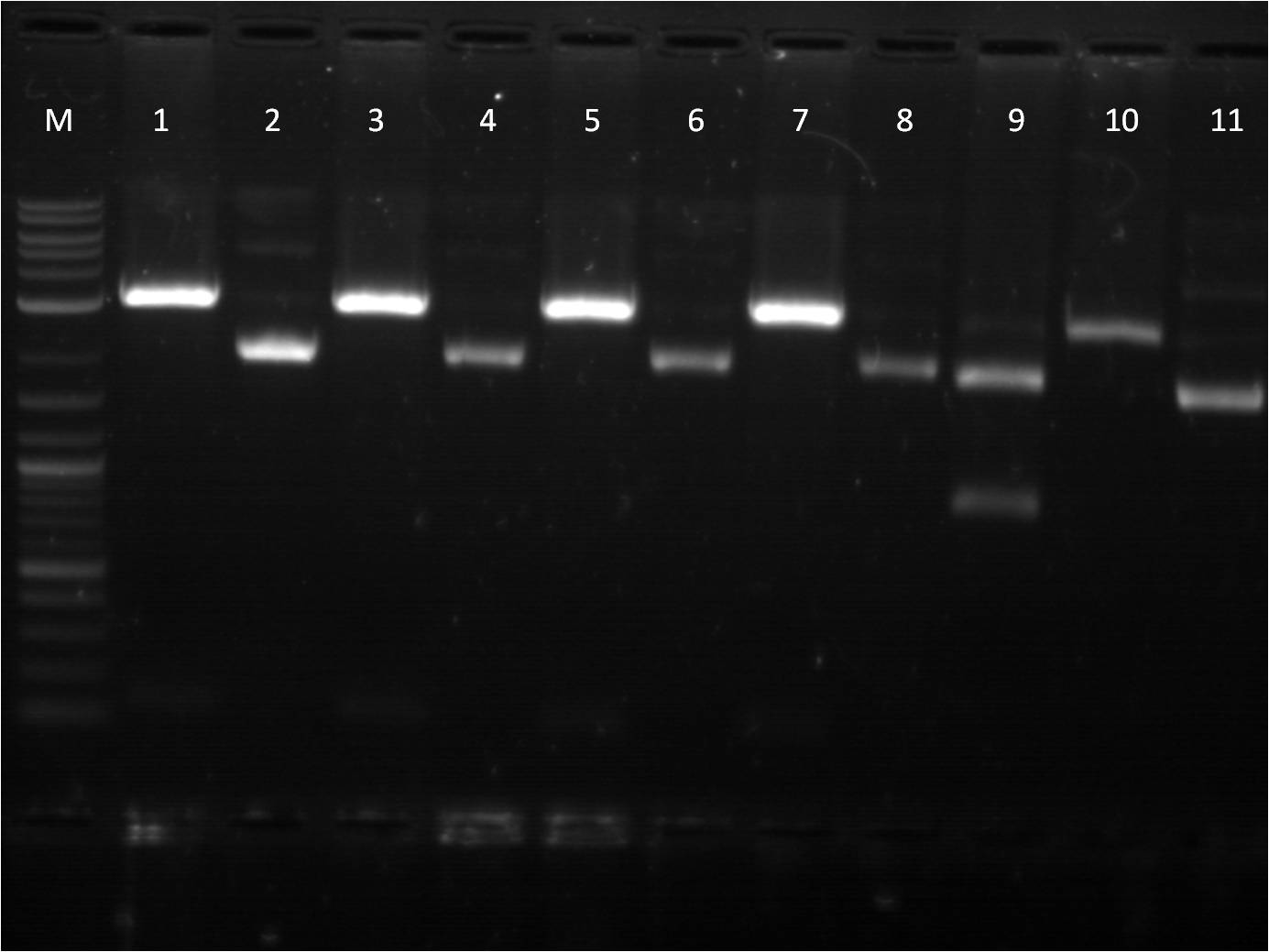
8/1: Today I set up the overnight cultures of my transformations from 7/28. I noticed that the RBS+RFP+term and RBS+BFP+Term both had a pinkish tint to it. This is fine for the RFP construct, but strange for the BFP construct. I plan on miniprepping the samples and then checking them on the gel. In the meantime, I did another round of ligations using a different RBS, J61101. We ran out of ampicillin plates, so only J61101+BFP+Term and J61101+RFP+Term were transformed and plated. 8/2: This morning I ran a gel of my RBS (J61100) +_FP's+term uncut plasmids to check their size. Ideally, RBS+GFP+Term should be ~3441 bp, RBS+RFP+Term ~2911 bp, and RBS+BFP+Term ~2940 bp.
M: ladder. 5: J61100+GFP+Term uncut 6: J61100+GFP+Term uncut 7: J61100+RFP+Term uncut 8: J61100+BFP+Term uncut 9: J61100 uncut On the gel, they seem to be the right base pair length. To further confirm that the plasmids are the correct construct, I am starting a method of restriction mapping. This technique is essentially doing a restriction digest and confirm the size of the insert that should be seen. In this case, I am cutting with XbaI and PstI because I can then use the insert for ligation with a promoter. I used our normal RD protocol. 8/3: This morning, I prepped samples for the QIAcube to run a miniprep protocol of our overnight cultures. Also, I set up a gel to run for my restriction mapping samples for J61100 + G/R/B_FP+term as well as my new RBS, J61101,+ R/B_FP +term constructs. M: Ladder. 1: J61100+GFP+Term A RD 2: J61100+GFP+Term B RD 3: J61100+RFP+Term RD 4: J61100+BFP+Term RD 5: J61100+RFP+Term uncut 6: J61100+BFP+Term uncut Here again, the restriction digest of the large construct does not seem to work. Only RBS+GFP+term A and B are seen to have two bands, yet they are at an incorrect length (should be 1362 bp rather than ~2500bp). The J61101+R/B_FP+Term seems to check out ok as far as expected base pair length. Going back to my problems with restriction digest from last week, I did another restriction digest with all 6 samples (4 J61100+_FP's+Term and 2 J61101+R/B_FP+Term) for a 4 hour incubation time since I am cutting with XbaI and PstI. M: Ladder. 8: J61100+GFP+Term A RD 9: J61100+GFP+Term B RD 10: J61100+RFP+Term RD 11: J61100+BFP+Term RD 12: J61101+RFP+Term RD |
WEEK 10: 8/8-8/12/2011 |
|
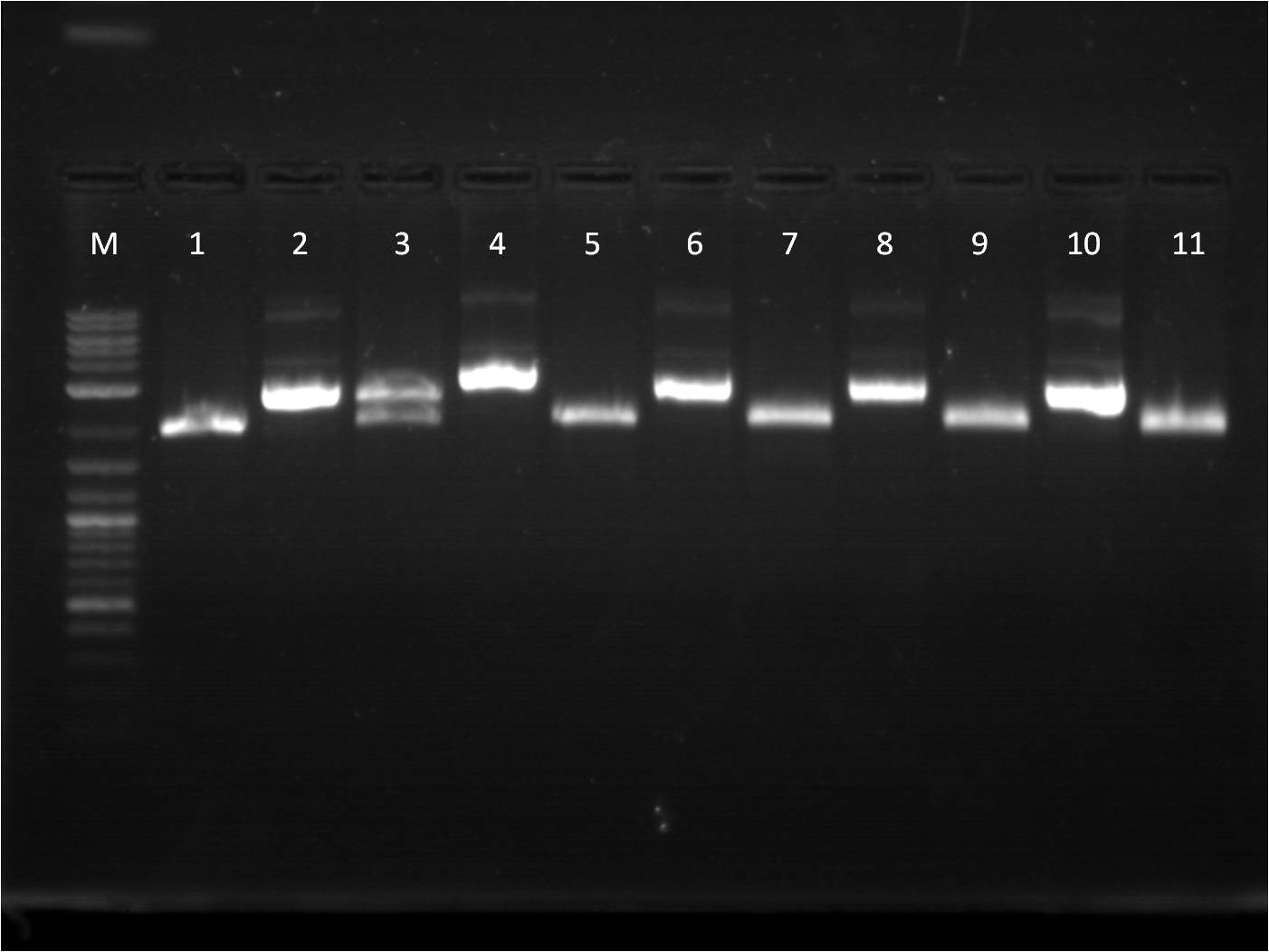
8/8: This week I am continuing to try to create a composite part for our RFP, GFP, and BFP reporters. I decided to use a more commmon RBS B0034 and ligate it with all 3 genes which already have a terminator attached. Also, I did a CIP reaction on J61100 and J61101 backbones which I then ligated with RFP and BFP. In addition to these "bottoms up" reporter parts, I am starting a new project of cloning the TetR gene which will inhibit our pTet promoter. I am using the biobrick part BBa_C0040. Luckily we already have a frozen stock of it, so the part can be spread-plated as well as prepared as an overnight culture right from the stock. 8/9: The transformations worked well, especially with the new RBS, B0034. Overnight cultures going in tonight. 8/10: Quantification of minipreps was decent, however when I did a restriction digest, cutting at X and P, only one band appeared. The uncut bands look around what they should be, but the fact is that I am not seeing an insert at all on the restriction digest reactions. Thus, I tried a controlled reaction, cutting with different combinations of enzymes that would still produce an expected insert. With the TetR part, C0040, I cut at E and S. M: Ladder. 1: B0034+RFP+Term RD 2: B0034+RFP+Term uncut 3: B0034+GFP+Term RD 4: B0034+GFP+Term uncut 5: J61100+BFP+Term RD 6: J61100+BFP+Term uncut 7: J61100+RFP+Term RD 8: J61100+RFP+Term uncut 9:J61101+RFP+Term RD 10: J61101+RFP+Term uncut 11: J61101+BFP+Term RD 8/11: The gel of the control reaction showed only single bands for the reporter parts. It confirms that the plasmids I currently have are incorrect as they don't contain the insert. Thus, I am going to start from scratch and use a two different methods of combinations to create the construct. I will try the original method I used first, which is to combine the fluorescent protein with the Terminator first. The other method is to combine the fluorescent protein to an RBS first (see diagram). Thus today, I will be setting up a restriction digest of RFP, GFP, and BFP cut for both methods. In terms of my TetR gene, C0040, it seems like there is also only one band showing and no insert. I will try the RD again, but using a larger amount of ng of DNA. insert DNA combination maps M: Ladder. 1: B0034+RFP+Term E,S 2: B0034+RFP+Term E,P 3: B0034+GFP+Term E,S 4: B0034+GFP+Term E,P 5: C0040.1 RD 6: C0040.1 uncut 7: C0040.2 RD 8: C0040.2 uncut 9:B0034+BFP+Term X,P 10: B0014+BFP+Term uncut
M: Ladder. 1: GFP X,P 2: GFP E,S 3: BFP X,P 4: BFP E,S 5: RFP X,P 6: RFP E,S 7: B0034 1A RD 8: B0034 1A uncut 9:B0034 1B 10: B0034 1B uncut |
WEEK 11: 8/14-8/19/2011 |
|
8/14: Came in on Sunday to transform my ligations of R/G/B_FP+term and R/G_FP+B0034. I also set up a restriction digest of P0440 (cutting at X,P) and I14033, pCat, (cutting at S,P). 8/15: In the morning I ran a gel of TetR composite and pCat RD's. Everything looks good, the DNA was extraction, I set up a ligation between them and transformed them. Transformations of the reporter genes looked good, overnight cultures were made. M: Ladder. 1: P0440.1 RD 2: P0440.1 uncut 3: P0440.2 RD 4: P0440.2 uncut 5: pCat.1 RD 6: pCat.2 RD 7: pCat uncut 8/16: The DNA quantification of the R/G/B_FP+term and R/G_FP+B0034 parts seemed good and I set up a restriction digest, cutting all with X,P. However, the gel did not come out as well. The R/G_FP+B0034 look correct, but the R/G/B_FP+term does not look right. It almost seems as if the fluorescent protein was never inserted and just terminator was there, given the insert band length. I extracted all of the parts anyways, but given the DNA quantification of the RFP and GFP + term, I only used BFP+term to ligate with an RBS. As for the R/G_FP+B0034, I combined this with pTet (R0040) as it is our strongest constitutive promoter. The ligation reactions were then transformed. M: Ladder. 1: RFP+Term RD 2: RFP+Term uncut 3: GFP+Term RD 4: GFP+Term uncut 5: BFP+Term RD 6: BFP+Term uncut 7: B0034+RFP RD 8: B0034+RFP uncut 9:B0034+GFP RD 10: B0034+GFP uncut
TOP M: Ladder. 1: BFP+Term.1 RD 2: BFP+Term.1 uncut 3: BFP+Term.2 RD 4: BFP+Term.2 uncut 5: BFP+Term.3 RD 6: BFP+Term.3 uncut 7: BFP+Term.4 RD 8: BFP+Term.4 uncut 9:pTet+GFPc X,P 10: pTet+GFPc E,X 11: pTet+GFPc uncut BOTTOM M: Ladder. 1: pTet+YFPc E,X 2: pTet+YFPc uncut 3: pTet+YFPc X,P 4: pTet+RFPc X,P 5: pTet+RFPc E,X 6: pTet+RFPc uncut 8/18: Today I ligated and transformed BFP and the CIP'd backbone, B0015 (Terminator). Our restock of SpeI enzyme just came today, so I did a restriction digest on my TetR+pCat (P0440+I14033), cutting at S,P and E,S so that I can both use it as an insert and backbone. M: Ladder. 5: pCat.1+TetR.1 S,P 6: pCat.1+TetR.1 uncut 7: pCat+TetR.3 E,S 8: pCat+TetR.3 uncut 9: pCat.1+TetR.1 uncut 10: pCat.1+TetR.3 uncut 11: pCat.3+TetR.1 uncut
8/20: Transformations from yesterday worked. However, the plates with the pTet+_FPc as a backbone were all glowing. The addition of the TetR gene to the construct should repress the pTet promoter. The plates with the TetR+pCat backbone had some colonies that glowed, but most did not. I also did a restriction digest of BFP+Term, cutting at X,P. |
WEEK 12: 8/22-8/26/2011 |
|
8/22: This morning I ran a gel of my BFP+Term RD's and pTet+B0034+G/RFP plasmids. The BFP+Term RD's looked great, having bands exactly where they should be. The uncut pTet+B0034+G/RFP plasmids looked about right, but I will have to do a restriction mapping of the parts to be sure. gel pic 8/22 I did a restriction digest of my pCat+TetR+pTet+R/YFPc and J23101+TetR plasmid to check if they are correct. I also did a restriction digest of K156026, which is RBS+BFP. I cut everything with X,P with the exception of J23101+TetR which was cut with E,S and S,P. The gel showed correct sizes for the pCat+TetR+pTet+R/YFPc as well as the RD of K156026. However, the bands of J23101+TetR were too faint to extract from the gel, but they appear to be correct. gel pic 8/22 Lastly, I did a ligation reaction of my BFP+Term parts with different RBS's (B0034, J61100, J61101, J61127). 8/23: After confirming the size of my composite plasmids, I had to make glycerol stocks of everything. I did another round of overnight culutres, minipreps, restriction digest, and gel electrophoresis to make sure the cell cultures I was making glycerol stocks of were correct. I made stocks of pCat+TetR+pTet+RFPc/GFPc/YFPc, pTet+B0034+GFP/RFP, BFP+Term, and pCat/J23101+TetR. All of these parts will be submitted to the Registry. Today, I also ligated K156026 with pTet and pCat and transformed all of my BFP ligaitons (RBS's+BFP+Term and K156026+pTet/pCat). 8/24: Today I did a restriction mapping of J23101+TetR, cutting with X,P, and made overnight cultures for K156026 and a part containing the backbone, pSB1C3, which we need all of our submitted parts in. The rest of the day was spent updating my notebook. 8/25: This afternoon I did miniprep's on all of the overnight cultures of my BFP transformations. In order to check all of the parts, I set up a restriction digest cutting all of the parts with X,P. This will both serve as a check to the size of the inserts and allow us to ligate the part with our desired backbone, pSB1C3. In anticipation of everything working, I made glycerol stocks of K156026, B0034+RFP/GFP, J61127/J61101/J61100+BFP+Term, and pCat/pTet+K156026. 8/26: Today is my last day of work. I ran a gel of my BFP composite part RD's as well as K156026 and B0034+RFP/GFP restriction digests. Everything looks correct size and these should all be good to go. gel pic 8/26
|
Resources
BioBricks
For access to all BioBrick parts and information, go to http://partsregistry.org/Main_Page
Lab Protocols
For a link to the protocols we use in our lab, go to Protocols
Tuberculosis Genes
For information on tuberculosis gene sequences and information, go to http://www.tbdb.org/
 "
"