File:UCL-content-GBS-theory-figure-1.jpg
From 2011.igem.org
No higher resolution available.
UCL-content-GBS-theory-figure-1.jpg (769 × 553 pixels, file size: 82 KB, MIME type: image/jpeg)
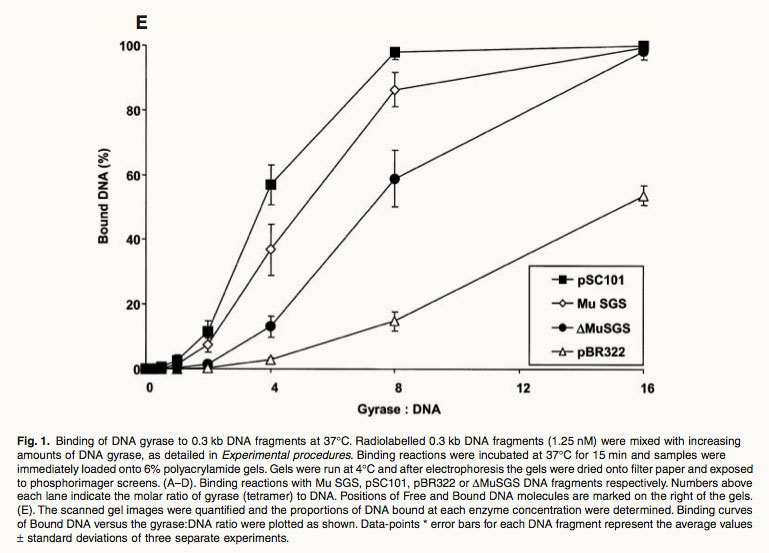
Fig. 1. Binding of DNA gyrase to 0.3 kb DNA fragments at 37∞C. Radiolabelled 0.3 kb DNA fragments (1.25 nM) were mixed with increasing amounts of DNA gyrase, as detailed in Experimental procedures. Binding reactions were incubated at 37∞C for 15 min and samples were immediately loaded onto 6% polyacrylamide gels. Gels were run at 4∞C and after electrophoresis the gels were dried onto filter paper and exposed to phosphorimager screens. (A–D). Binding reactions with Mu SGS, pSC101, pBR322 or DMuSGS DNA fragments respectively. Numbers above each lane indicate the molar ratio of gyrase (tetramer) to DNA. Positions of Free and Bound DNA molecules are marked on the right of the gels. (E). The scanned gel images were quantified and the proportions of DNA bound at each enzyme concentration were determined. Binding curves of Bound DNA versus the gyrase:DNA ratio were plotted as shown. Data-points * error bars for each DNA fragment represent the average values ± standard deviations of three separate experiments. Ref: Oram, M., et al., A biochemical analysis of the interaction of DNA gyrase with the bacteriophage Mu, pSC101 and pBR322 strong gyrase sites: the role of DNA sequence in modulating gyrase supercoiling and biological activity. Mol Microbiol, 2003. 50(1): p. 333-47.
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 04:43, 22 September 2011 |  | 769×553 (82 KB) | PhilippBoeing (Talk | contribs) | |
| 11:01, 21 September 2011 |  | 769×553 (113 KB) | Eintisar (Talk | contribs) | (Fig. 1. Binding of DNA gyrase to 0.3 kb DNA fragments at 37∞C. Radiolabelled 0.3 kb DNA fragments (1.25 nM) were mixed with increasing amounts of DNA gyrase, as detailed in Experimental procedures. Binding reactions were incubated at 37∞C for 15 min a) |
File links
The following page links to this file:
 "
"