Team:uOttawa/Results
From 2011.igem.org
Results
BioBrick characterization
Characterization of BBa_K642000 and BBa_K642004
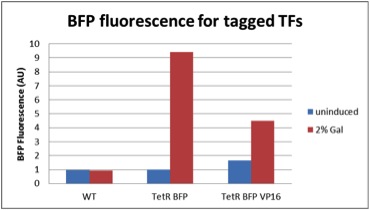
We quantified the fluorescence of two tagged repressors: BBa_K642000 and BBa_K642004 by cloning them downstream of the yeast Gal10/1 promoter. The construct was then integrated using KanMX6 selection into the Ade2 locus of a BY4742 strain of S. cerevisiae. The resulting strain was innoculated in 3 mL of 1x SM + 2% Galactose + 2% Adenine overnight. The overnight culture was reinoculated into 1 mL of the same media at an OD600 of 0.02. After three hours the strain was analyzed for BFP fluorescence on a Cyan ADP flow cytometer.
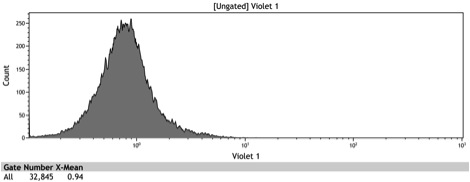
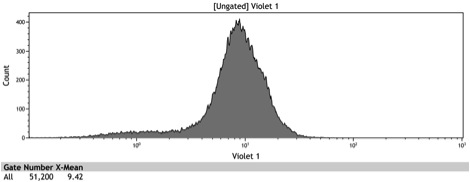
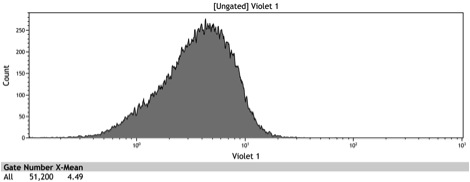
Figure 1: In vivo BFP fluorescence of BBa_K642000, a TetR repressor tagged with yeast codon optimized BFP and BBa_K642004, a TetR repressor tagged with yeast codon optimized BFP and VP16 activation domain. These parts were integrated using KanMX6 selection into a BY4742 strain of S. cerevisiae.
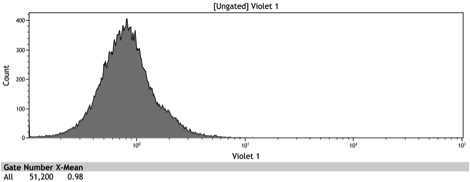
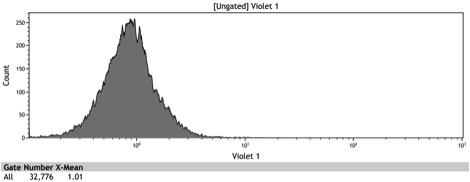
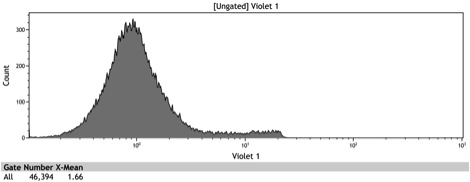
Figure 2: Histogram of uninduced wildtype BY4742
Figure 3: Histogram of uninduced BBa_K642000 -TetR-BFP
Figure 4: Histogram of uninduced BBa_K642004 - TetR-BFP-VP16
Figure 5: Histogram of induced wildtype BY4742
Figure 6: Histogram of induced BBa_K642000 – TetR-BFP
Figure 7: Histogram of induced BBa_K642004 – TetR-BFP-VP16
 "
"