Team:Washington/Magnetosomes/Results
From 2011.igem.org
(→Magnetosome Toolkit:) |
|||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| - | ===Magnetosome Toolkit:=== | + | ===About the Magnetosome Toolkit:=== |
[[File:Washington Methode image.jpg|center|955px]] | [[File:Washington Methode image.jpg|center|955px]] | ||
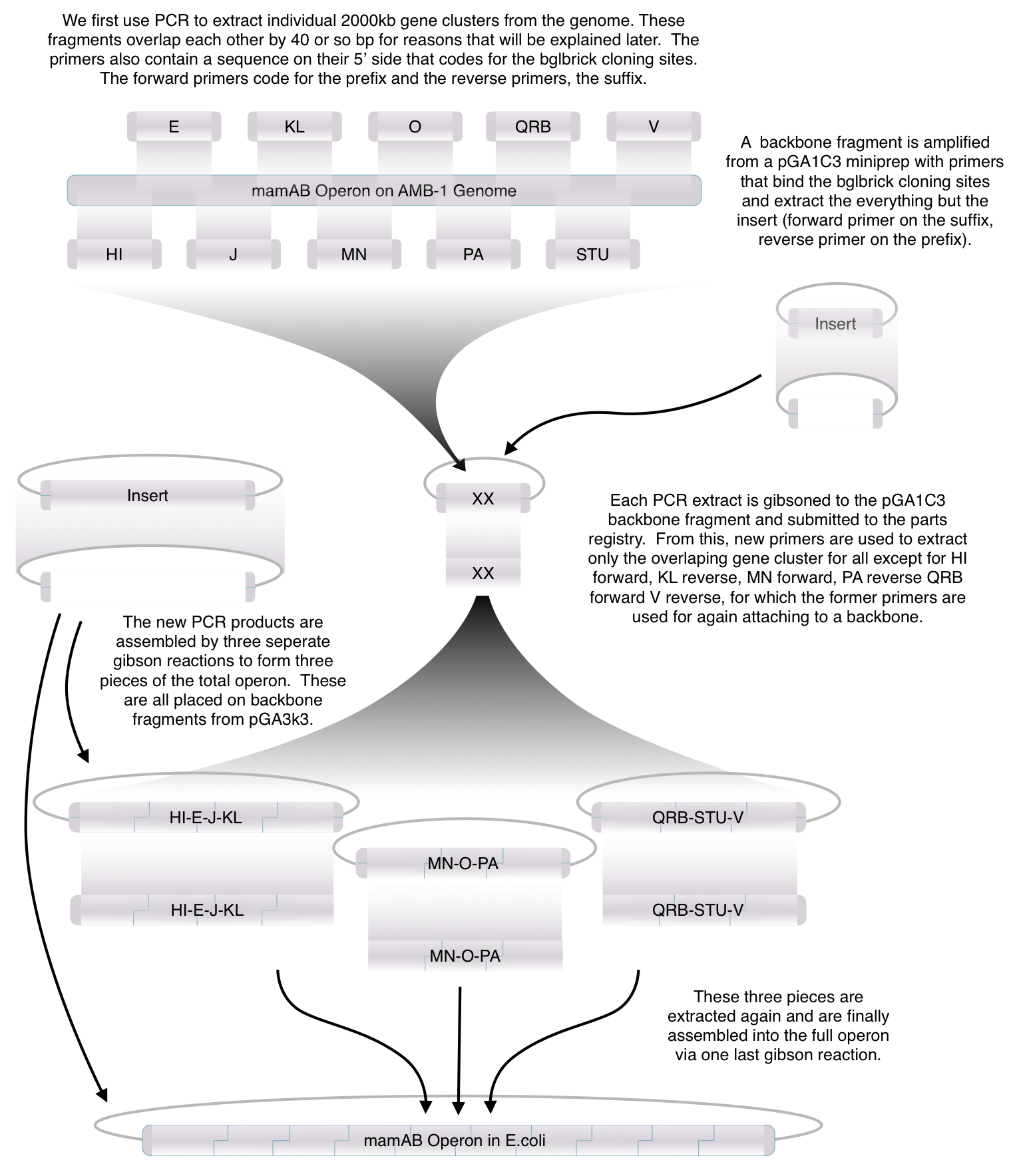
| - | Using the vectors we created in our Gibson Assembly Toolkit, our team was able to create a "Magnetosome Toolkit" consisting of the most basic parts required for future | + | Using standard synthetic biology protocols and the vectors we created in our Gibson Assembly Toolkit, our team was able to create a '''"Magnetosome Toolkit"''' consisting of the most basic parts required for magnetosome formation. Providing this toolkit allows future iGem teams to manipulate and understand magnetosome formation to one day create magnets in various types of bacteria. |
| - | + | ===What' in the toolkit and How we assembled it=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | These genes were visualized on gels and sequence confirmed. However, in order to make sure that gene-activity had not be lost, we created mam gene-sfGFP fusions, transformed them into E.coli, and imaged them with microscopy. The two most notable constructs were mamK-sfGFP and mamI-sfGFP | + | |
| + | After piecing together the 16 kb genome of the mamAB gene cluster within the magnetosome island (MAI), we extracted out the genes in the following group: | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Gene groups | ||
| + | ! Length (bp) | ||
| + | |- | ||
| + | | mamHI | ||
| + | | 1541 | ||
| + | |- | ||
| + | | mamE | ||
| + | | 2172 | ||
| + | |- | ||
| + | | mamJ | ||
| + | | 1538 | ||
| + | |- | ||
| + | | mamKL | ||
| + | | 1336 | ||
| + | |- | ||
| + | | mamMN | ||
| + | | 2323 | ||
| + | |- | ||
| + | | mamO | ||
| + | | 1914 | ||
| + | |- | ||
| + | | mamPA | ||
| + | | 1493 | ||
| + | |- | ||
| + | | mamQRB | ||
| + | | 2029 | ||
| + | |- | ||
| + | | mamSTU | ||
| + | | 2030 | ||
| + | |- | ||
| + | | mamV | ||
| + | | 1002 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | Using standard PCR protocols, these genes were extracted and..... | ||
| + | These genes were visualized on gels and sequence confirmed; therefore, we can be sure that these are some of the many genes required for proper magnetosome formation. | ||
| + | |||
| + | |||
| + | |||
| + | However, in order to make sure that gene-activity had not be lost, we created mam gene-sfGFP fusions, transformed them into E.coli, and imaged them with microscopy. The two most notable constructs were mamK-sfGFP and mamI-sfGFP | ||
(--> we're going to put pictures here....<--) | (--> we're going to put pictures here....<--) | ||
Revision as of 20:09, 16 September 2011
About the Magnetosome Toolkit:
Using standard synthetic biology protocols and the vectors we created in our Gibson Assembly Toolkit, our team was able to create a "Magnetosome Toolkit" consisting of the most basic parts required for magnetosome formation. Providing this toolkit allows future iGem teams to manipulate and understand magnetosome formation to one day create magnets in various types of bacteria.
What' in the toolkit and How we assembled it
After piecing together the 16 kb genome of the mamAB gene cluster within the magnetosome island (MAI), we extracted out the genes in the following group:
| Gene groups | Length (bp) |
|---|---|
| mamHI | 1541 |
| mamE | 2172 |
| mamJ | 1538 |
| mamKL | 1336 |
| mamMN | 2323 |
| mamO | 1914 |
| mamPA | 1493 |
| mamQRB | 2029 |
| mamSTU | 2030 |
| mamV | 1002 |
Using standard PCR protocols, these genes were extracted and..... These genes were visualized on gels and sequence confirmed; therefore, we can be sure that these are some of the many genes required for proper magnetosome formation.
However, in order to make sure that gene-activity had not be lost, we created mam gene-sfGFP fusions, transformed them into E.coli, and imaged them with microscopy. The two most notable constructs were mamK-sfGFP and mamI-sfGFP
(--> we're going to put pictures here....<--)
-essential gene a picture of gel, notable constructs, process diagram, and cool results please.
 "
"