Team:Washington/Celiacs/Results
From 2011.igem.org
(→=Using a whole cell lysate assay to screen a large number of mutants for good activity) |
(→Testing our designed mutants for activity on PQLP) |
||
| Line 8: | Line 8: | ||
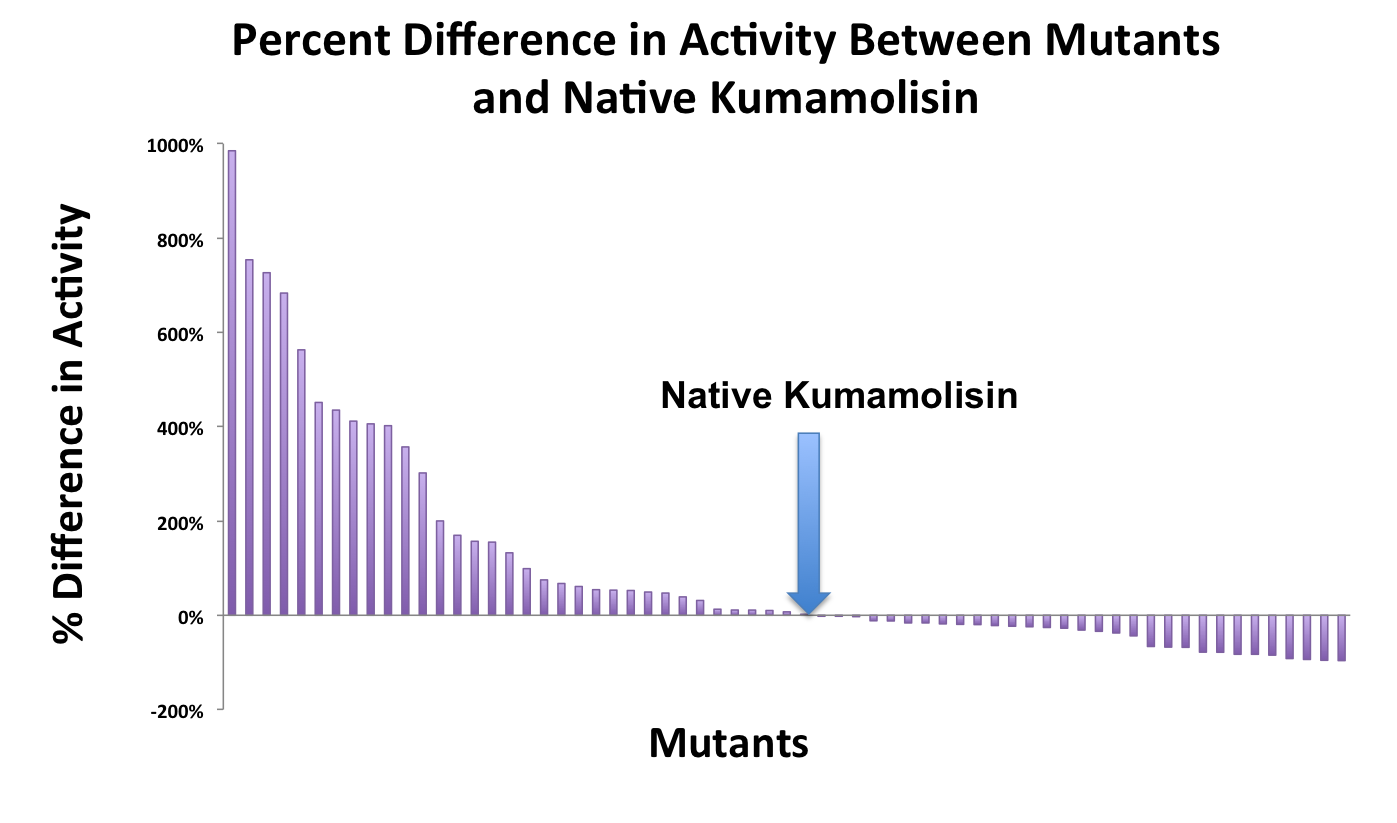
In order to determine whether our proposed mutations to the wild-type Kumamolisin improved the ability of the enzyme to break down PQLP, we tested each mutant with a whole cell lysate fluorescence assay. Cells harboring the expressed mutants were lysed at pH 4, mimicking the gastric environment. The released enzymes, after being roughly separated from cell material, were added to a fluorescent PQLP that had been conjugated to a quencher. Thus, no fluorescence was achieved until the peptide had been cleaved and the fluorophore had been released from the quencher. This allowed a relative assessment of rate of enzyme activity by measuring increase in fluorescence of the system. | In order to determine whether our proposed mutations to the wild-type Kumamolisin improved the ability of the enzyme to break down PQLP, we tested each mutant with a whole cell lysate fluorescence assay. Cells harboring the expressed mutants were lysed at pH 4, mimicking the gastric environment. The released enzymes, after being roughly separated from cell material, were added to a fluorescent PQLP that had been conjugated to a quencher. Thus, no fluorescence was achieved until the peptide had been cleaved and the fluorophore had been released from the quencher. This allowed a relative assessment of rate of enzyme activity by measuring increase in fluorescence of the system. | ||
| - | As one might expect, our first screen of mutants showed some mutants with a decrease in activity from the wild-type, some showed no change, and some actually showed great increase in activity. One single point mutant showed close to a 1000% increase | + | As one might expect, our first screen of mutants showed some mutants with a decrease in activity from the wild-type, some showed no change, and some actually showed great increase in activity. One single point mutant showed close to a 1000% increase in activity from wild-type Kumamolisin! |
[[File:Washington Mutant Screen Percent IncDec.jpg|left|942px|thumb|Over 100 unique mutants were screened with a whole cell lysate assay for improved activity on the PQLP model substrate.]] | [[File:Washington Mutant Screen Percent IncDec.jpg|left|942px|thumb|Over 100 unique mutants were screened with a whole cell lysate assay for improved activity on the PQLP model substrate.]] | ||
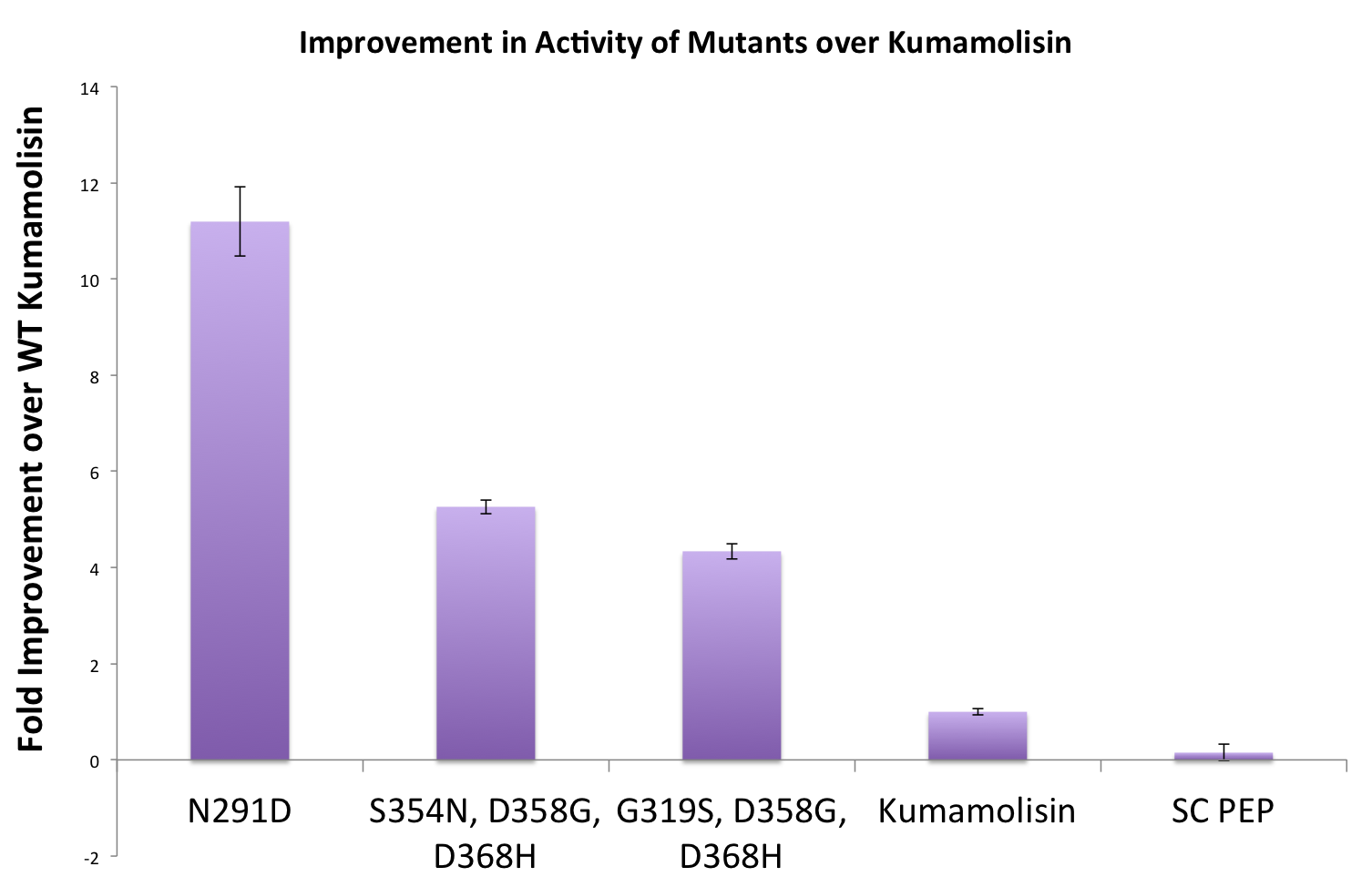
| + | Once we had identified mutants that showed a promising increase in activity from the wild-type kumamolisin, we purified and characterized activity in concentration controlled fluorescence assays, identical to the fluorescence system used for the whole cell lysate assay. Our best mutant demonstrated an 11-fold increase in activity from the native enzyme. | ||
| - | |||
[[File:Washington Best Mutants.png|left|500px|thumb|Our final engineered enzyme showed activity over 100 fold higher than wild type Kumamolisin, and ~700 fold higher than SC-PEP.]] | [[File:Washington Best Mutants.png|left|500px|thumb|Our final engineered enzyme showed activity over 100 fold higher than wild type Kumamolisin, and ~700 fold higher than SC-PEP.]] | ||
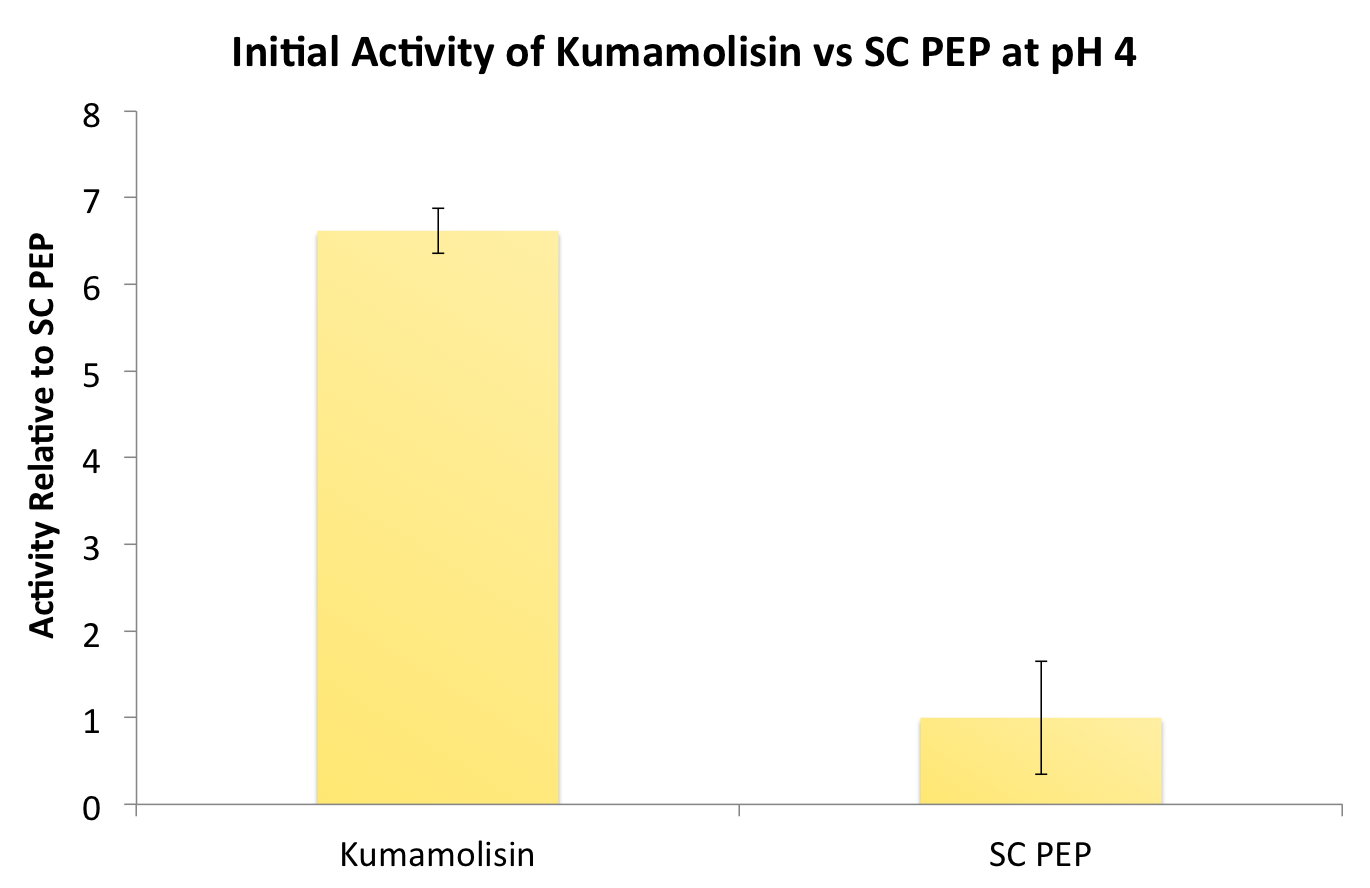
[[File:Washington InitialKumavSC.png|left|500px|thumb|Our final engineered enzyme showed activity over 100 fold higher than wild type Kumamolisin, and ~700 fold higher than SC-PEP.]] | [[File:Washington InitialKumavSC.png|left|500px|thumb|Our final engineered enzyme showed activity over 100 fold higher than wild type Kumamolisin, and ~700 fold higher than SC-PEP.]] | ||
Revision as of 02:05, 16 September 2011
Testing our designed mutants for activity on PQLP
Using a whole cell lysate assay to screen a large number of mutants for good activity
In order to determine whether our proposed mutations to the wild-type Kumamolisin improved the ability of the enzyme to break down PQLP, we tested each mutant with a whole cell lysate fluorescence assay. Cells harboring the expressed mutants were lysed at pH 4, mimicking the gastric environment. The released enzymes, after being roughly separated from cell material, were added to a fluorescent PQLP that had been conjugated to a quencher. Thus, no fluorescence was achieved until the peptide had been cleaved and the fluorophore had been released from the quencher. This allowed a relative assessment of rate of enzyme activity by measuring increase in fluorescence of the system.
As one might expect, our first screen of mutants showed some mutants with a decrease in activity from the wild-type, some showed no change, and some actually showed great increase in activity. One single point mutant showed close to a 1000% increase in activity from wild-type Kumamolisin!
Once we had identified mutants that showed a promising increase in activity from the wild-type kumamolisin, we purified and characterized activity in concentration controlled fluorescence assays, identical to the fluorescence system used for the whole cell lysate assay. Our best mutant demonstrated an 11-fold increase in activity from the native enzyme.
 "
"