Team:Washington/Alkanes/Future
From 2011.igem.org
(Difference between revisions)
| (37 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| - | : | + | <center><big><big><big><big>'''Diesel Production: Future Directions'''</big></big></big></big></center><br><br> |
| - | [https://2011.igem.org/Team:Washington/Alkanes/Future/Vector System Optimization] | + | Our current ''in vivo'' alkane production system efficiently makes C15 alkanes. However, to be efficient enough for factory production, there are two broad goals to be done: |
| + | # Increase production efficiency | ||
| + | # Increase the diversity of the range of alkanes that can be produced. | ||
| + | # Increase the scale of the system for industrial processes. | ||
| + | We have already begun efforts to expand the efficiency and scope of alkane production, '''check them out below!''' | ||
| + | |||
| + | [[Image:UW_2011_Alkane_Future_Work_Image.png|7px||right]] | ||
| + | |||
| + | [[Image:Washington system optimization.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/Vector]] | ||
| + | <br><br> | ||
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/Vector '''System Optimization'''] | ||
:<nowiki> The efficiency of this system was reported to be much higher than our initial system, but the growth conditions of the assay and the DNA was not available to us. We increased production efficiency by altering the initial environmental conditions in the assay.</nowiki> | :<nowiki> The efficiency of this system was reported to be much higher than our initial system, but the growth conditions of the assay and the DNA was not available to us. We increased production efficiency by altering the initial environmental conditions in the assay.</nowiki> | ||
| + | <br><br> | ||
| - | [https://2011.igem.org/Team:Washington/Alkanes/Future/DecarbDesign Decarbonylase Redesign] | + | [[Image:Washington_2011_ADC_Redesign.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/DecarbDesign]] |
| - | :<nowiki>One way to diversify the kind of alkanes produced is to alter the substrate specificity of the proteins involved. | + | <br><br> |
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/DecarbDesign '''Decarbonylase Redesign'''] | ||
| + | :<nowiki>One way to diversify the kind of alkanes produced is to alter the substrate specificity of the proteins involved. Since an alternative alkane-producing enzyme has not been found, we decided to mutate the aldehyde decarbonylase to produce shorter-chain alkanes.</nowiki> | ||
| + | <br><br> | ||
| - | [https://2011.igem.org/Team:Washington/Alkanes/Future/LuxCDE | + | [[Image:Washington_2011_LuxCDE.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/LuxCDE]] |
| - | :<nowiki>Another way to diversify our system is to use alternative proteins. Our current system uses acyl-ACP reductase, and we've identified an hypothetical alternative system that produces aldehydes: LuxCDE, made from parts from the 2010 </nowiki> | + | <br> |
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/LuxCDE '''Alternative Aldehyde'''] | ||
| + | :<nowiki>Another way to diversify our system is to use alternative proteins. Our current system uses acyl-ACP reductase, and we've identified an hypothetical alternative system that produces aldehydes: LuxCDE, made from parts from the 2010 competition.</nowiki> | ||
| + | <br><br><br> | ||
| - | [https://2011.igem.org/Team:Washington/Alkanes/Future/FabH2 | + | [[Image:Washington_2011_fabh2_branch.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/FabH2]] |
| - | :<nowiki> | + | <br> |
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/FabH2 '''Alternitive Alkane Products'''] | ||
| + | :<nowiki> The PetroBrick is only capable of producing unbranched, odd chain length alkanes, as the cell mainly utilizes straight chained, even chain length fatty acids. However, fuel we use is composed of a wide range of products. By changing which fatty acids are made by alkane producing cells, we could theoretically change which alkanes are being produced by the PetroBrick. </nowiki> | ||
| + | <br> | ||
| - | [https://2011.igem.org/Team:Washington/Alkanes/Future/Localization Enzyme Localization] | + | [[Image:Washington 2011 Protein Localization.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/Localization]] |
| - | :<nowiki> The easiest way to increase yield is to increase the concentration; if the number of molecules remained the same but the volume decreased, the reaction speed increases with | + | <br> |
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/Localization '''Enzyme Localization'''] | ||
| + | :<nowiki> The easiest way to increase yield is to increase the concentration; if the number of molecules remained the same but the volume decreased, the reaction speed increases with occurrences of molecular collisions. However, this is only easily done with purified protein assays. However, there are ways to "increase the concentration" of reactions in cells. We cannot make the cells literally denser, but we can localize the enzymes involved in the reaction, which either decreases the volume of the cell in which the enzymes reside and/or limit the number of reaction steps.</nowiki> | ||
| - | [https://2011.igem.org/Team:Washington/Alkanes/Future/Yeast Alternative | + | [[Image:Washington_2011_Alternate_Chassis.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/Yeast]] |
| + | <br> | ||
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/Yeast '''Alternative Chassis'''] | ||
:<nowiki> By synthesizing alkanes in organisms, alkanes can become a useful renewable energy source; CO2 emitted from burning the alkanes are converted to glucose through photosynthesis, and then the glucose can be fed to alkane-synthesizing organisms to restart the cycle. This cycle can be drastically improved if there were fewer steps in the cycle. We explored the possibility of optimizing the alkane biosynthesis system in different micro-organisms, with the idea that perhaps with autotrophic organisms, we do not need to obtain glucose for the reaction.</nowiki> | :<nowiki> By synthesizing alkanes in organisms, alkanes can become a useful renewable energy source; CO2 emitted from burning the alkanes are converted to glucose through photosynthesis, and then the glucose can be fed to alkane-synthesizing organisms to restart the cycle. This cycle can be drastically improved if there were fewer steps in the cycle. We explored the possibility of optimizing the alkane biosynthesis system in different micro-organisms, with the idea that perhaps with autotrophic organisms, we do not need to obtain glucose for the reaction.</nowiki> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | [[Image:Washington_FB.png|140px||left|link=https://2011.igem.org/Team:Washington/Alkanes/Future/Modeling]] | |
| + | <br> | ||
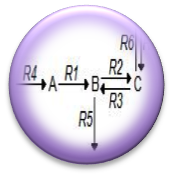
| + | [https://2011.igem.org/Team:Washington/Alkanes/Future/Modeling '''System Modeling'''] | ||
| + | :<nowiki> We have begun Flux Balance Modeling the alkane production system in order to better understand the carbon flux through the pathway. We believe that by understanding the curent flux through the pathway we can intelligently reengineer the platform in order to maximize flux to alkanes. </nowiki> | ||
Latest revision as of 00:35, 29 October 2011
Our current in vivo alkane production system efficiently makes C15 alkanes. However, to be efficient enough for factory production, there are two broad goals to be done:
- Increase production efficiency
- Increase the diversity of the range of alkanes that can be produced.
- Increase the scale of the system for industrial processes.
We have already begun efforts to expand the efficiency and scope of alkane production, check them out below!
- The efficiency of this system was reported to be much higher than our initial system, but the growth conditions of the assay and the DNA was not available to us. We increased production efficiency by altering the initial environmental conditions in the assay.
- One way to diversify the kind of alkanes produced is to alter the substrate specificity of the proteins involved. Since an alternative alkane-producing enzyme has not been found, we decided to mutate the aldehyde decarbonylase to produce shorter-chain alkanes.
- Another way to diversify our system is to use alternative proteins. Our current system uses acyl-ACP reductase, and we've identified an hypothetical alternative system that produces aldehydes: LuxCDE, made from parts from the 2010 competition.
- The PetroBrick is only capable of producing unbranched, odd chain length alkanes, as the cell mainly utilizes straight chained, even chain length fatty acids. However, fuel we use is composed of a wide range of products. By changing which fatty acids are made by alkane producing cells, we could theoretically change which alkanes are being produced by the PetroBrick.
- The easiest way to increase yield is to increase the concentration; if the number of molecules remained the same but the volume decreased, the reaction speed increases with occurrences of molecular collisions. However, this is only easily done with purified protein assays. However, there are ways to "increase the concentration" of reactions in cells. We cannot make the cells literally denser, but we can localize the enzymes involved in the reaction, which either decreases the volume of the cell in which the enzymes reside and/or limit the number of reaction steps.
- By synthesizing alkanes in organisms, alkanes can become a useful renewable energy source; CO2 emitted from burning the alkanes are converted to glucose through photosynthesis, and then the glucose can be fed to alkane-synthesizing organisms to restart the cycle. This cycle can be drastically improved if there were fewer steps in the cycle. We explored the possibility of optimizing the alkane biosynthesis system in different micro-organisms, with the idea that perhaps with autotrophic organisms, we do not need to obtain glucose for the reaction.
- We have begun Flux Balance Modeling the alkane production system in order to better understand the carbon flux through the pathway. We believe that by understanding the curent flux through the pathway we can intelligently reengineer the platform in order to maximize flux to alkanes.
 "
"