Team:WashU/Project
From 2011.igem.org
(→Results) |
(→Results) |
||
| Line 55: | Line 55: | ||
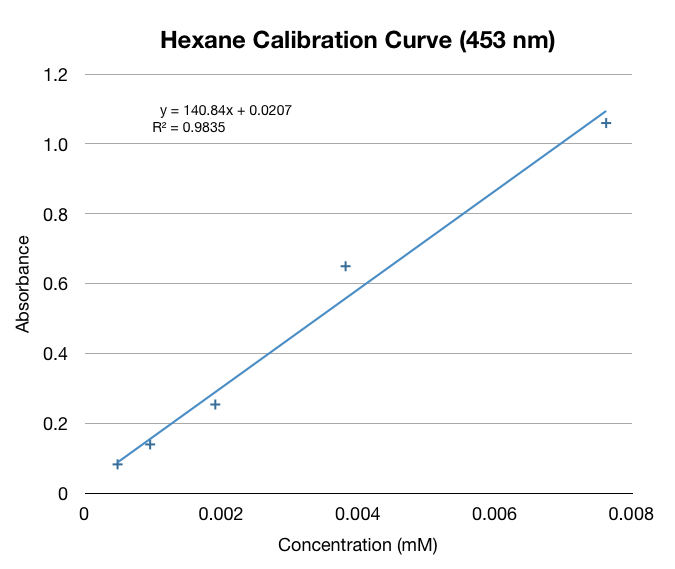
With our procedure and results we were able to derive formulas that correlated absorbance from the spectrophotometer to the total beta-carotene/beta-ionone concentration in yeast cells per gram dry weight of yeast cells. | With our procedure and results we were able to derive formulas that correlated absorbance from the spectrophotometer to the total beta-carotene/beta-ionone concentration in yeast cells per gram dry weight of yeast cells. | ||
| - | [[File:Formula.jpg]] | + | [[File:Formula.jpg|center]] |
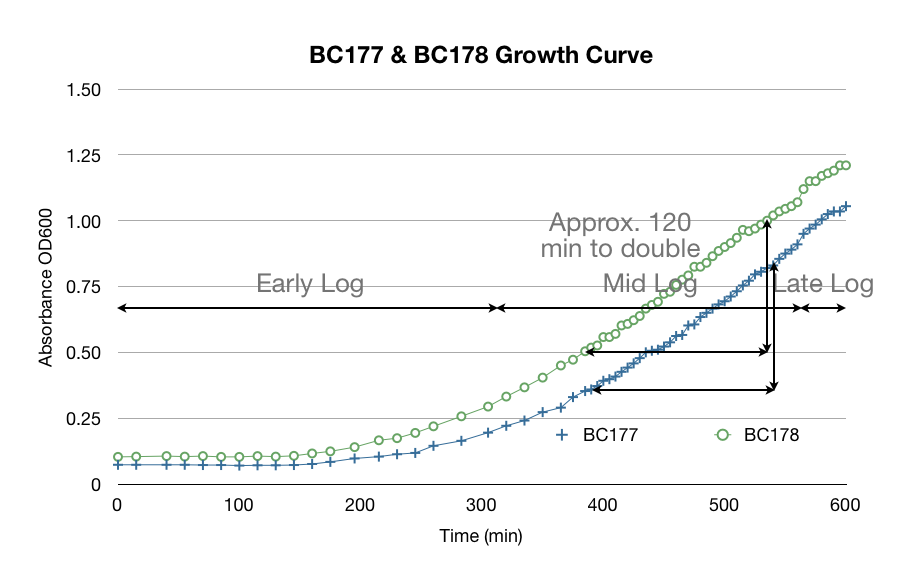
[[File:Yeast Growth Curve.jpg]] | [[File:Yeast Growth Curve.jpg]] | ||
Revision as of 22:41, 27 September 2011
Abstract
Engineering Carotenoid Biosynthesis in Saccharomyces cerevisiae
Vitamin A deficiency causes blindness in over 250,000 children annually. The WashU iGEM team hopes to address this issue by creating a transgenic strain of Saccharomyces cerevisiae (baker's yeast) that produces beta-carotene, the precursor to vitamin A. The WashU team created four DNA constructs for homologous recombination into the S. cerevisiae genome that will catalyze the production of beta-carotene. Each construct consisted of a gene encoding an enzyme from Xanthophyllomyces dendrorhous, a bacterium that produces beta-carotene. Three of the constructs encode for enzymes in the metabolic pathway required for beta-carotene production, while a fourth enzyme cleaves beta-carotene to form beta-ionone, a rose-scented compound used in the fragrance industry. Additionally, the WashU team has established spectraphotometric assays to detect beta-carotene and beta-ionone in yeast extract. Although we have yet to successfully incorporate these four genes into S. cerevisiae, we have prepared all four constructs and biobricked these genes for future use.
Background
This year, the WashU team has decided to study the enzymatic pathway responsible for the biosynthesis of B-Carotene and B-Ionone.
Our project, which includes the making of recombinant yeast that produce B-carotene and B-Ionone, is entirely based off the article High-Level Production of Beta-Carotene in Saccharomyces cerevisiae by Successive Transformation with Carotenogenic Genes from Xanthophyllomyces dendrorhous published in Applied and Environmental Microbiology. This paper explains the enzymes required for the B-carotene enzymatic pathway as well as the efficiency of the pathway when certain enzymes are missing or added. This paper can be found here.
Our goal this summer is to recreate this pathway by inserting the crtYB, crtI, and crtE genes into the yeast genome. Additionally, we would like to take this pathway one step further by adding the CCD1 gene which cleaves B-carotene to produce B-ionone. Both of these compounds have the potential to be commercially relevant to the yeast-production industry. Transgenic yeast that produces B-carotene could theoretically be used in the same manner that baker's yeast is used currently. When the GMO yeast is added to bread or other baked goods, it would produce B-carotene in addition to its normal byproducts which would then be infused in the bread. When consumed by the human body, B-carotene is automatically converted to vitamin A. GMO yeast that produces B-ionone also has the potential to be commerically viable. B-ionone is a fragrant chemical that is characterized by a rose scent and is widely used by the perfume industry. Currently, B-ionone is produced by organic synthesis; however, biosynthesis has the potential to provide an alternative production method for this useful compound.
During the process of creating transgenic yeast that produce B-Carotene and B-Ionone, we plan to BioBrick all four of genes involved in the pathway.
Project Goals
Results
Team WashU succeeded in creating expression constructs for each of the four genes in the carotenoid biosynthesis pathway. Subsequently, we have submitted eight original biobricked constructs in the "DNA Planning" stage in the Registry of Standard Biological Parts. Despite the fact that we have all of our DNA sequences complete and synthsized, we ran out of time before we could transform our yeast strains or submit our DNA constructs to the Registry of Standard Biological Parts. However, should next year's WashU IGEM team decide to continue this project, the eight biobricks we submitted would undoubtedly be completed for use by future IGEM teams.
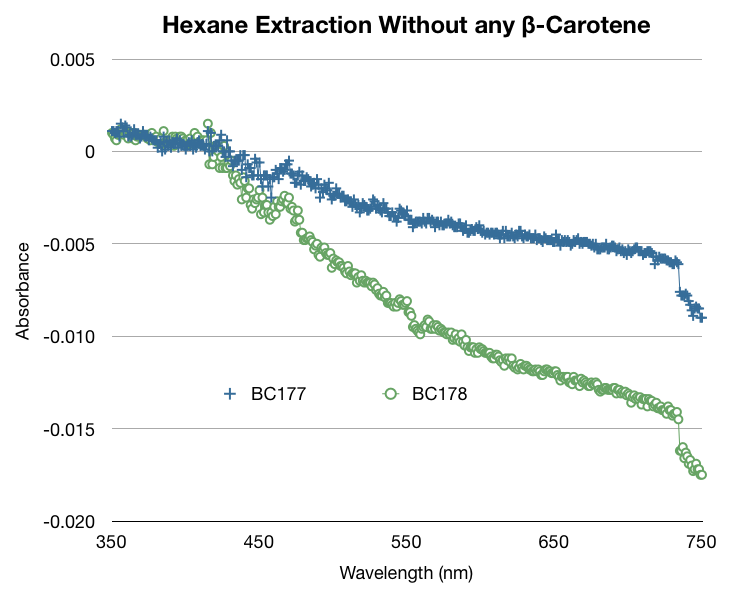
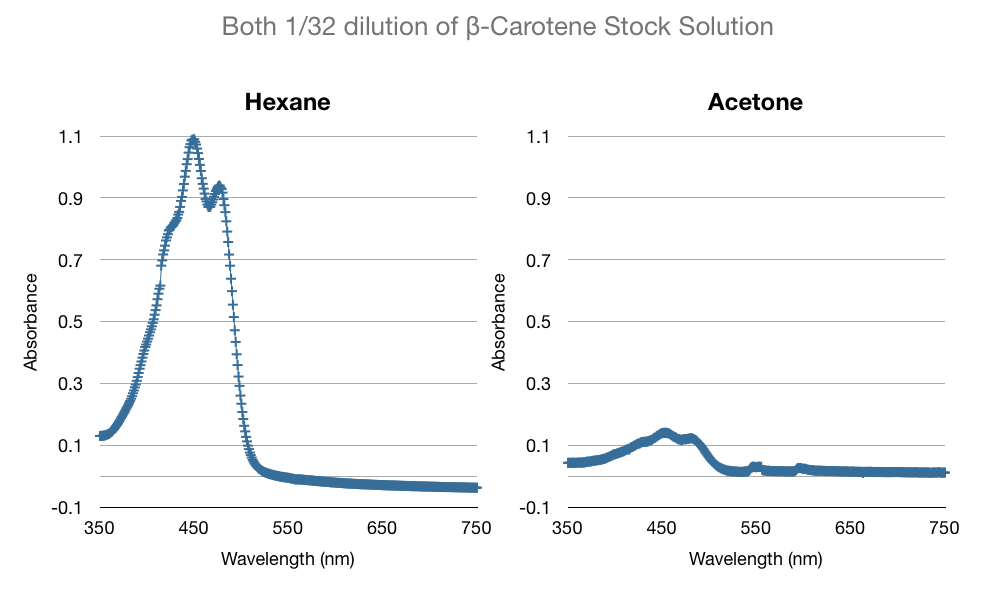
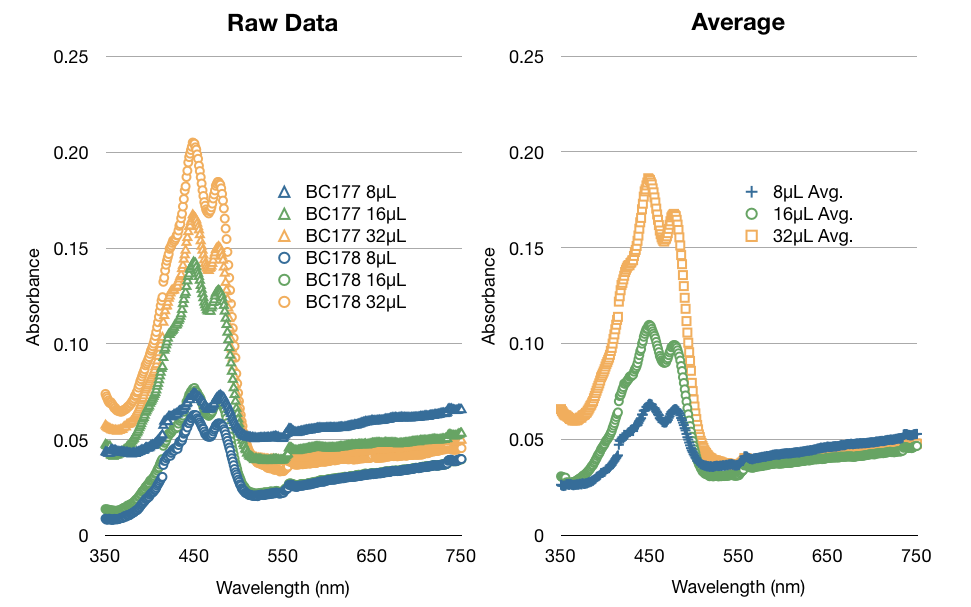
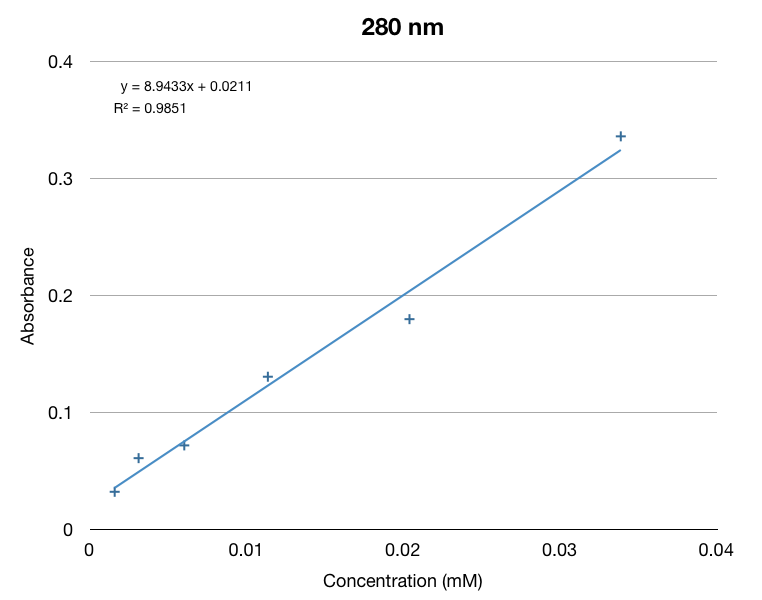
In addition to our work regarding the creation of recombinant yeast, Team WashU carried out many assays for the quantitative detection of beta-carotene and beta-ionone in yeast extract. This data and assay procedure can be found below.
We based our assay procedure from "Carotenoid Analysis" procedure from “High-Level Production of Beta-Carotene in Saccharomyces cerevisiae...” Verwaal et. al (2007)
We made several modifications to the procedure to meet our needs, yeast strains, beta carotene/beta ionone concentrations, and efficiency expectations.
1. Pellet yeast cells by centrifuging for 5 minutes at 10,000g.
2. Discard supernatant. Resuspend cells in 1mL sterile H2O. Vortex for 10 sec.
3. Add 1 g 0.50-0.75mm glass beads. Vortex for 5 min to break cells.
4. Add 2.5 mL of 0.2% (wt/vol) pyrogallol dissolved in methanol. Vortex cells for 10 sec.
5. Add 1.25 mL 60% (wt/vol) KOH. Vortex cells for 10 sec.
6. Incubate cells for 1 hr at 75°C. Vortex every 15 min for saponification.
7. Add 2 mL of hexane to extract the carotenoid fraction. Vortex for 1 min.
8. Centrifuge tubes for 5 min at 10,000g.
9. Mix hexane layer by pipetting up and down.
10. Pipette 1.2mL of hexane layer out into sealed test tube.
11. Pipette 1 mL of hexane into cuvette.
12. Measure absorption on spectrophotometer.
With our procedure and results we were able to derive formulas that correlated absorbance from the spectrophotometer to the total beta-carotene/beta-ionone concentration in yeast cells per gram dry weight of yeast cells.
 "
"