Project Description
1. Introduction
The aim of this project is to design and implement a system exhibiting sustained oscillatory protein expression which is synchronized across a population of spatially constrained E. coli cells. The principles that govern this type of behavior have been studied both in theory and in practice, and as such there exists a solid foundation to apply these ideas in the context of the iGEM competition. In essence, this project consists of constructing a plasmid which contains protein encoding genes which reciprocally affect each other’s expression in a reliable manner, and experimentally measuring the expression dynamics to test the predictive value of a mathematical model. Due to the specificity of the required system parameters, and resulting difficulty in experimentally verifying the phenomena we wish to observe, special considerations regarding the experimental set-up had to be made. We hope that this system might be employed as a pace-making device to drive more complex genetic circuits requiring time-dependent gene expression, or as a component in sophisticated metabolic engineering applications.
Fig.1. Artistic rendering of the Synchronized Oscillatory System.
2. Mechanism
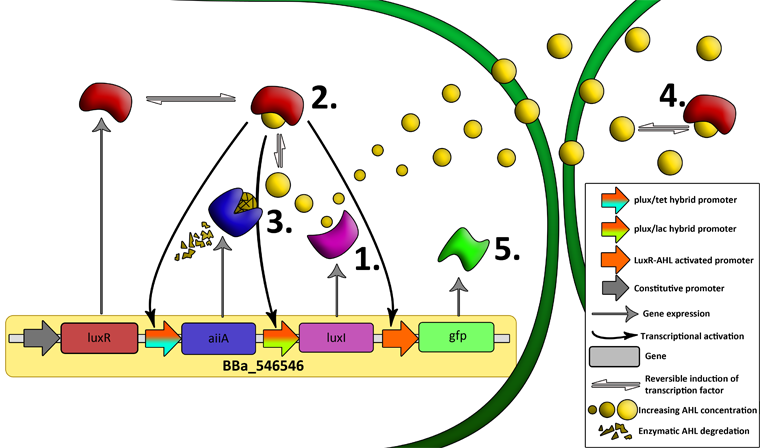
There are a number of genetic circuit topologies that have the potential to exhibit oscillatory behavior under the right conditions. However, the requirement that the oscillations should be synchronized posed a constraint on the components that could be used. The starting point for our genetic circuitry was a design recently published in the article “A synchronized quorum of genetic clocks” by Danino et al. This design combines elements of the Vibrio fischeri quorum sensing system with a quorum quenching enzyme from Bacillus subtilis, resulting in coupled positive and negative feedback loops which regulate the expression of a reporter protein.
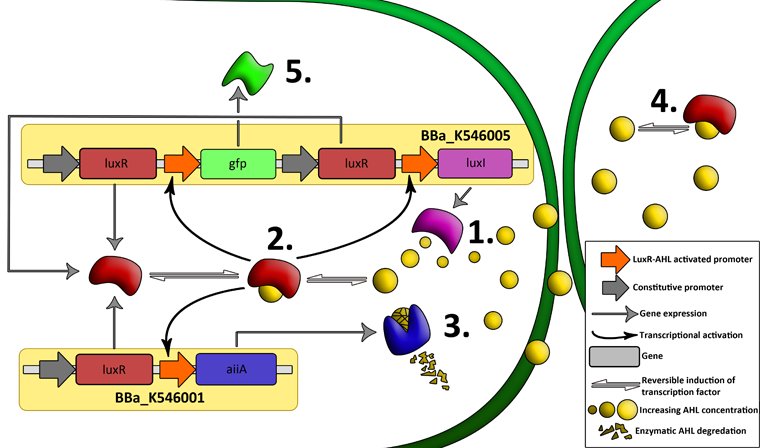
Fig.2. Genetic circuit of a synchronized oscillator
Basic Components:
LuxR is a transcriptional regulator in the bioluminescent quorum-sensing system of the symbiotic deep sea bacterium Vibrio fischeri. It is induced by binding the auto-inducer molecule N-(3-oxohexanoyl)-homoserine lactone (AHL). The AHL-LuxR complex controls expression of the lux regulon, which contains diverging pRight and pLeft promoter elements. The pRight element has low basal transcription, and is activated by AHL-LuxR; pLeft has higher basal expression, and is repressed by the AHL-LuxR complex. This dual activity makes LuxR a useful element for controlling interconnected genetic feedback loops. The unrestricted diffusion of AHL through the plasma membrane allows spatially proximate populations of cells to experience identical AHL conditions and synchronize AHL-dependent gene expression.
The enzyme LuxI is an acyl-homoserine-lactone synthase which produces the intercellular signalling molecule N-(3-oxohexanoyl)-homoserine lactone (AHL). Placing LuxI under control of the pRight promoter results in a positive feedback loop: when increases in cell density cause the intracellular AHL concentration to rise above the activation threshold of the pRight promoter, the transcription rate of the LuxI gene is increased which in turn results in the production of more AHL.
AiiA is an enzyme from B. subtilis which degrades AHL. Its biological function is to interfere with the quorum sensing signals of other bacteria. Placing it under control of the pRight promoter results in negative feedback as a response to increasing AHL concentrations. The reporter molecule Green Fluorescent Protein (GFP) is also regulated by the pRight promoter and provides a quantitative (albeit delayed) indication of the AHL concentration the cell is exposed to at a given point in time. LuxI, AiiA and GFP are all tagged for rapid degradation (LVA-tag). Due to differences in the synthesis and degradation rates of LuxI and AiiA, there exists a space of conditions within which periodic oscillations in AHL concentration, and concomitant oscillatory protein expression can emerge. Under most conditions, the level of AHL within a population of cells will quickly reach a steady state. However, by simulating the system using a quantitative biochemical model, it is possible to predict conditions under which oscillations are likely to occur. See our modeling page for details.
3. Designs
“Hasty” system:
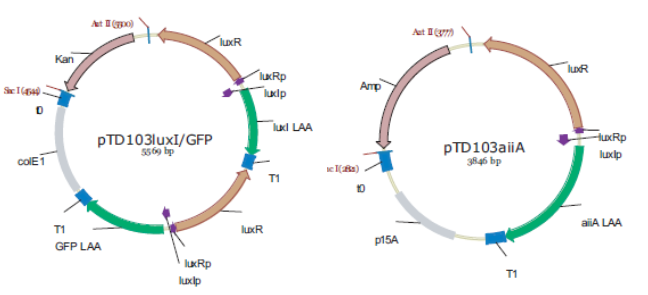
Fig.4. Synchronized Oscillator plasmids using natural quorum sensing system (Danino et al. 2010)
The first design we implemented was a BioBrick part based on reconstruction of the plasmids used by Danino et al. We intended to make as accurate a replica as possible in order to confirm the previously published results, and to test the viability of our experimental platform. However, there are a few differences between the original Hasty system and “our replica“. While the Hasty system employs the natural lux promoter which contains divergent pLeft and pRight elements (Fig 4), the BioBrick parts we employed have both elements in the same orientation. Both the original and our system contain 3 copies of the luxR gene under control of the pLeft element. Furthermore, a different (high copy) backbone was used during the functional validation of the parts, as opposed to the low copy backbone employed by Hasty.
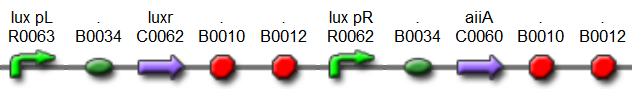
 Fig.5. BioBrick part K546005 forms the positive feedback loop component of the basic synchonized oscillator.
Fig.5. BioBrick part K546005 forms the positive feedback loop component of the basic synchonized oscillator.
Fig.6. BioBrick part K546001 forms the negative feedback loop component and complements K546005.
Due to the tendency of interconnected positive and negative feedback loops to reach a steady state rather than sustaining oscillations, the external conditions need to be precisely controlled in order for the system to produce synchronized oscillations. In this system, the only parameters relevant to oscillatory gene expression that can be controlled during operation are the cell density and the external AHL degradation rate (see model for details).
However, maintaining a constant cell density over extended periods of time while independently varying the flow rate (AHL degradation) is difficult to achieve using traditional cultivation and measurement set-ups. In order to control these parameters while keeping the cells microscope-accessible, we developed a custom fluidic platform that can trap cells and allow them to remain viable over the duration of an experiment while maintaining a constant and reproducible cell density (See flow chamber page for details).
Double Tunable Synchronized Oscillator
The aforementioned constraints are due to the fact that there are no means by which the expression kinetics of LuxI and AiiA can be externally influenced. In order to gain more control over the expression dynamics, we refactored the system and introduced “tuning knobs” in the form of chemically inducible hybrid promoters (pR/LacI inducible by IPTG and pR/tetR inducible by anhydrotetracycline). The addition of two independent control variables substantially expands the parameter space (cell density, flow rate) within which oscillations can occur. We also made the design more streamlined by taking advantage of the great diversity of available BioBrick quorum sensing parts in order to remove redundant elements and fit the entire system onto a single plasmid (a 30% reduction in size compared to the original design). Note: in order to take advantage of the tuning capabilities it is necessary to co-transform a plasmid which constitutively expresses LacI or TetR, or both.
Fig.6. Genetic circuit of a synchronized oscillator
 Fig.7. The complete synchronized oscillator circuit with a single copy of constitutively expressed LuxR
Fig.7. The complete synchronized oscillator circuit with a single copy of constitutively expressed LuxR
4. Experimental verification
The individual subcomponents of the oscillatory circuit were tested using a fluorescence photospectrometer. Changes in GFP expression relative to different AHL concentrations were measured to verify the functionality of the parts (see data page). However, due to the precise growth conditions required for synchronized oscillations to be sustained, it was necessary to adopt a more sophisticated approach to test the complete system.
There were three requirements for the experimental set-up to test the synchronized oscillator system:
a) ability to grow cells in a physically constrained space to achieve the high cell density necessary for quorum sensing machinery to respond. Ideally, the cell density should remain more or less constant over the course of hours, in addition to being predictable and reproducible.
b) supplying cells with fresh media to provide nutrients and remove waste (and AHL) without affecting the cell density.
c) directly observe cells with a fluorescence microscope to perform time-lapse studies aiming at detecting changes in GFP expression over a period of hours.
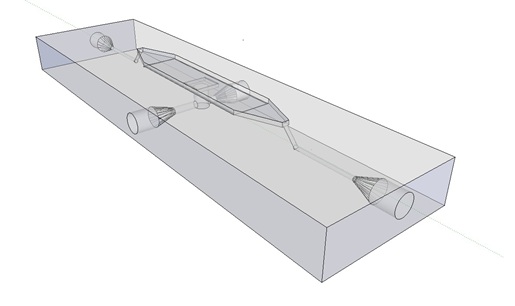
Previous studies of oscillatory gene expression have employed microfluidic devices with micron-scale trapping chambers to restrict cells. However, these systems can be prohibitively expensive, require specialized accessories, and are generally not reuseable. We developed a low-cost experimental flow chamber that fulfills all of the aforementioned criteria (for detailed specifications see flow chamber page) .
Fig.8. 3D rendering of the flow chamber.
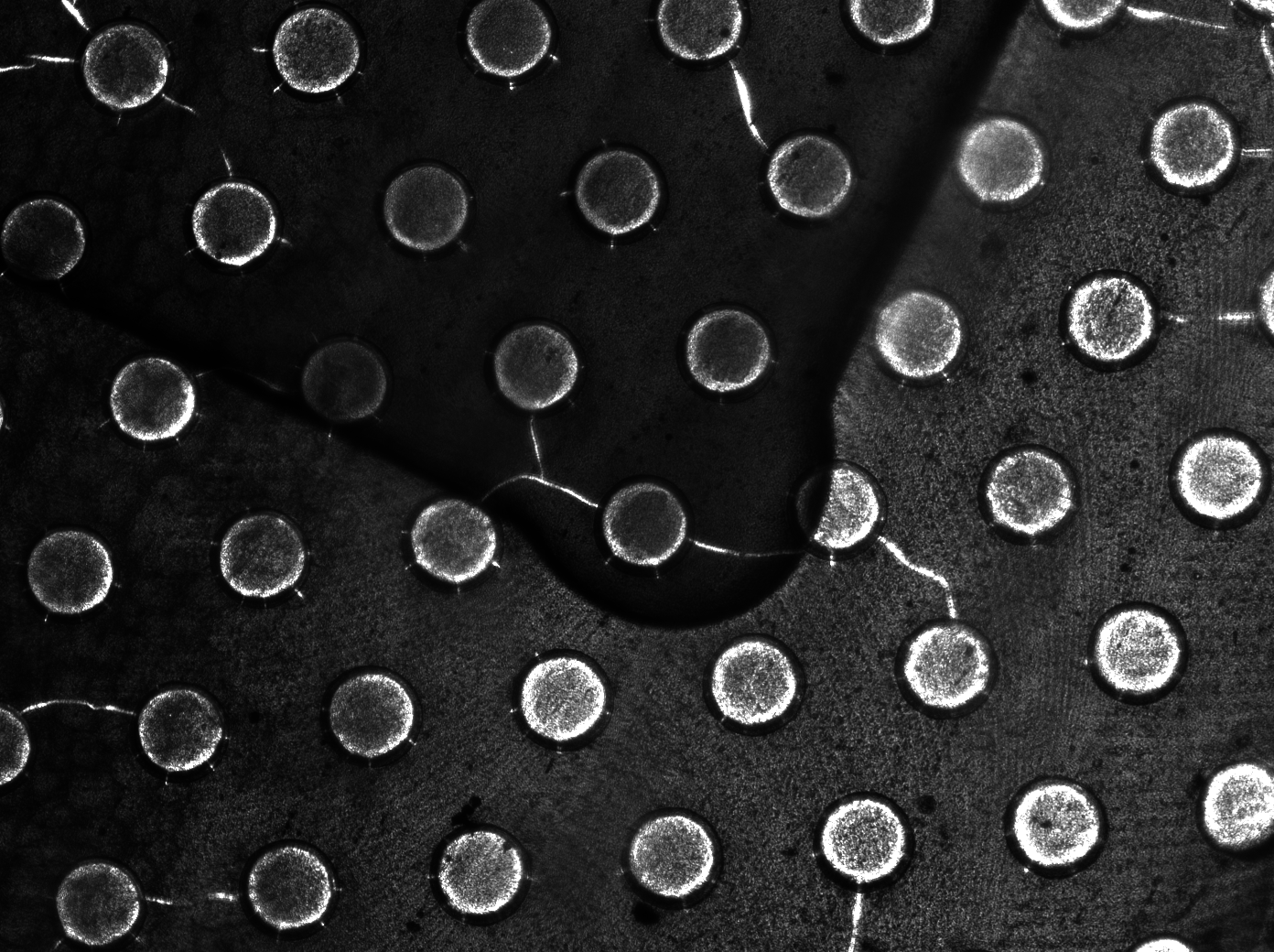
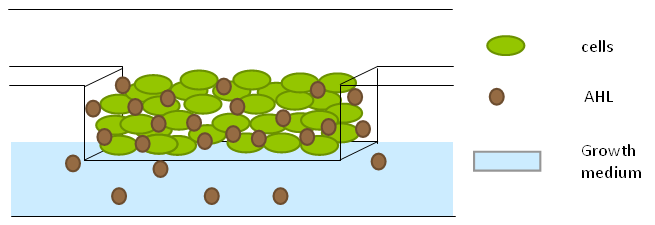
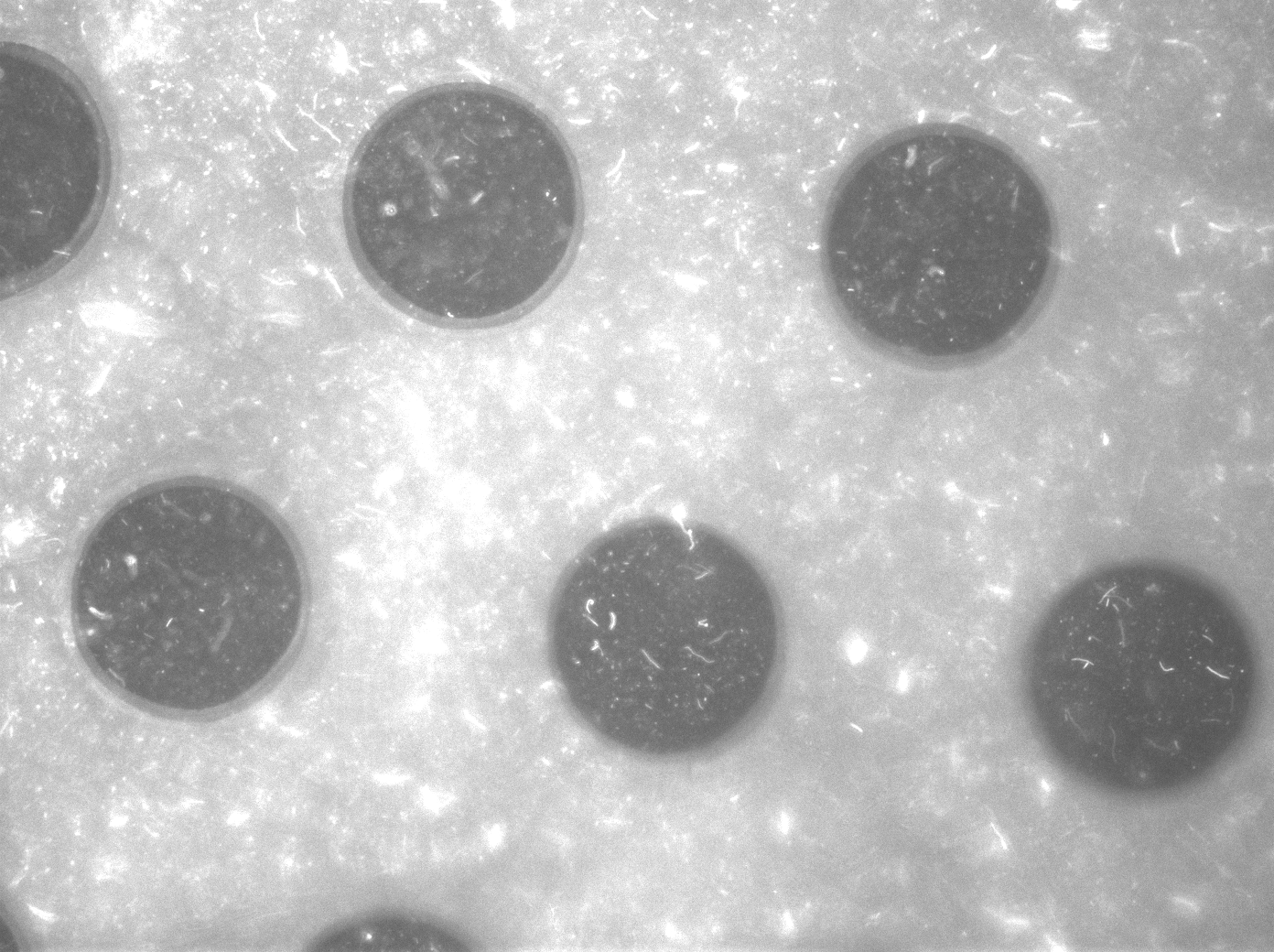
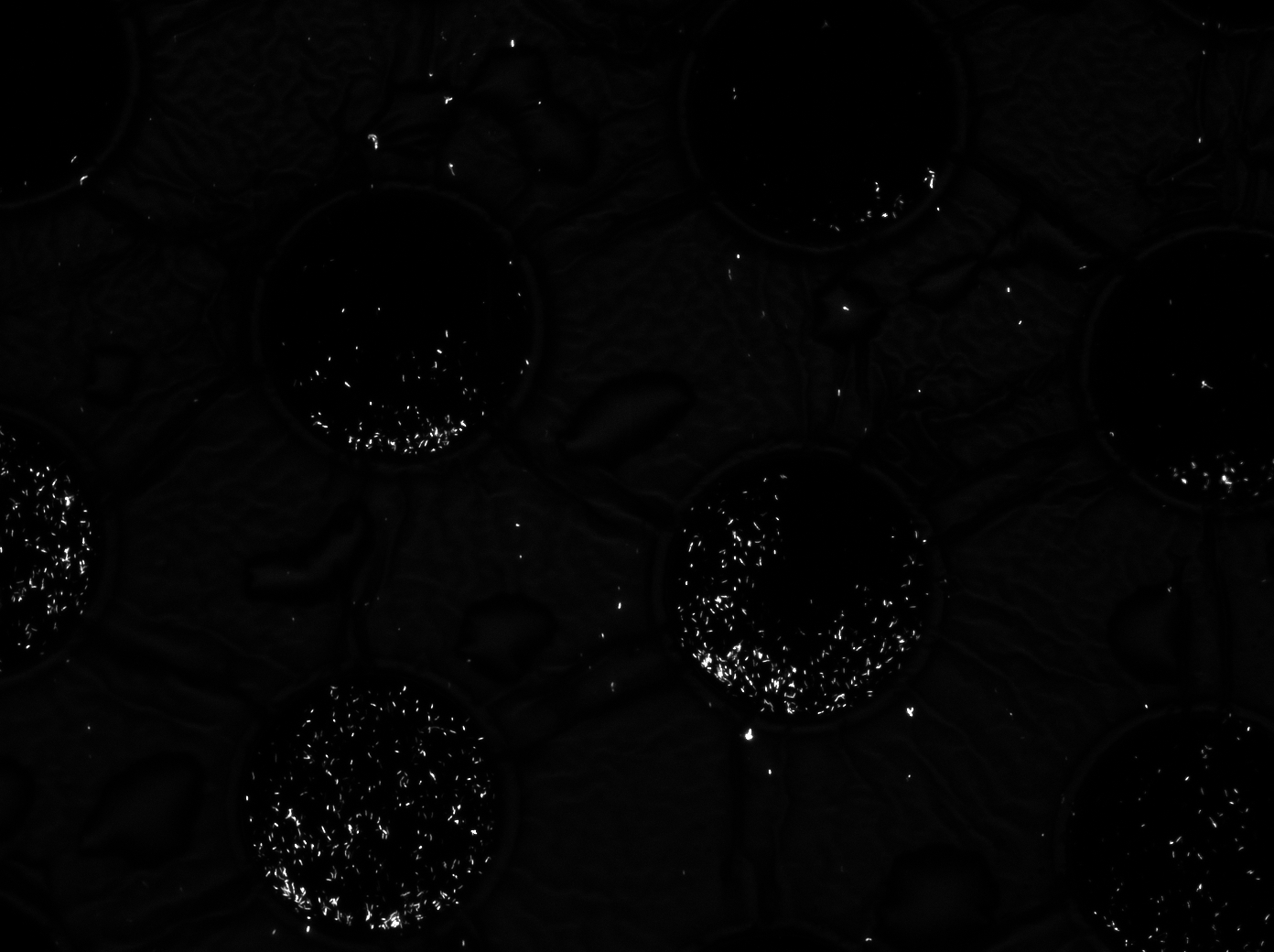
To measure the oscillations, the flow chamber was outfitted with a micro-dish, which is a thin aluminium oxide sheet containing thousands of circular wells. The wells are 180 μm in diameter and 40 μm deep. Small molecules (such as nutrients from growth media) can diffuse through the bottom of the dish and feed cells growing in the wells. For these experiments, the cells were fed through bottom in order to restrict their growth to the wells. This allowed the cells to reach high densities and relative stable populations which could be studied individually (5 wells could be visualized using 100x magnification)
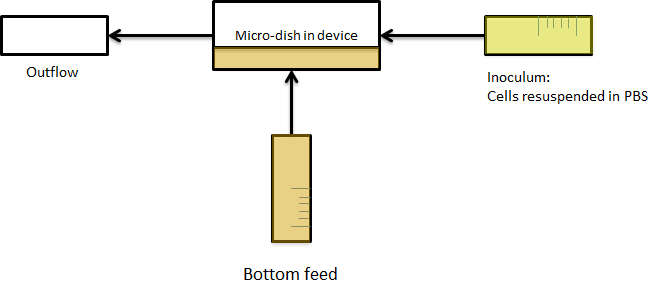
File:Dish.gifFig.9. Above: Schematic illustration of the micro-dish.
Fig.10. Right: Microdish installed in flow chamber viewed at 40x magnification, illuminated from the bottom with white light. Growth medium is supplied from the bottom through the 5x5mm socket (rounded corner). Diffusion of cells through the aluminium oxide allows cells to grow across the entire dish.
The cells containing the synchronized oscillator system plasmid were grown overnight in LB medium supplemented with an antibiotic. They were then centrifuged for 10 minutes at 4600 rcf and resuspended in Phosphate Buffered Saline solution before being injected through the top flow port (Fig.11). Directly after injection, the cells fill the entire chamber volume (Fig.12). After approximately 5-8 hours the cells situated outside the wells are starved no longer express detectable levels of GFP (Fig.13). Cells in the wells continue to grow for approximately 12 hours before reaching a stable cell density (Fig.14). After the wells were saturated with cells (Fig.15), time lapse microscopy was initiated. A photograph was taken every 10 minutes using a fluorescence microscope fitted with a GFP filter. Changes in GFP expression over timewere subsequently quantified. (Video 1)
 "
"