Team:WHU-China/Standard
From 2011.igem.org
Golden Gate Method
In the development of synthetic biology, the standardized biobrick is a classic, ingenious and delicate design. Thanks to the standardized biobrick, researchers can conveniently build biological regulatory network through the classic splicing method. However, under the condition of actual operation, some regulatory systems are so complicated that classic splicing method may not work as well: too many biobricks are needed in the assembly, which lead to tedious experimental process and increasing demand of time. As to this problem, WHU-China team make an attempt to apply golden gate method to the assembly of biobricks.
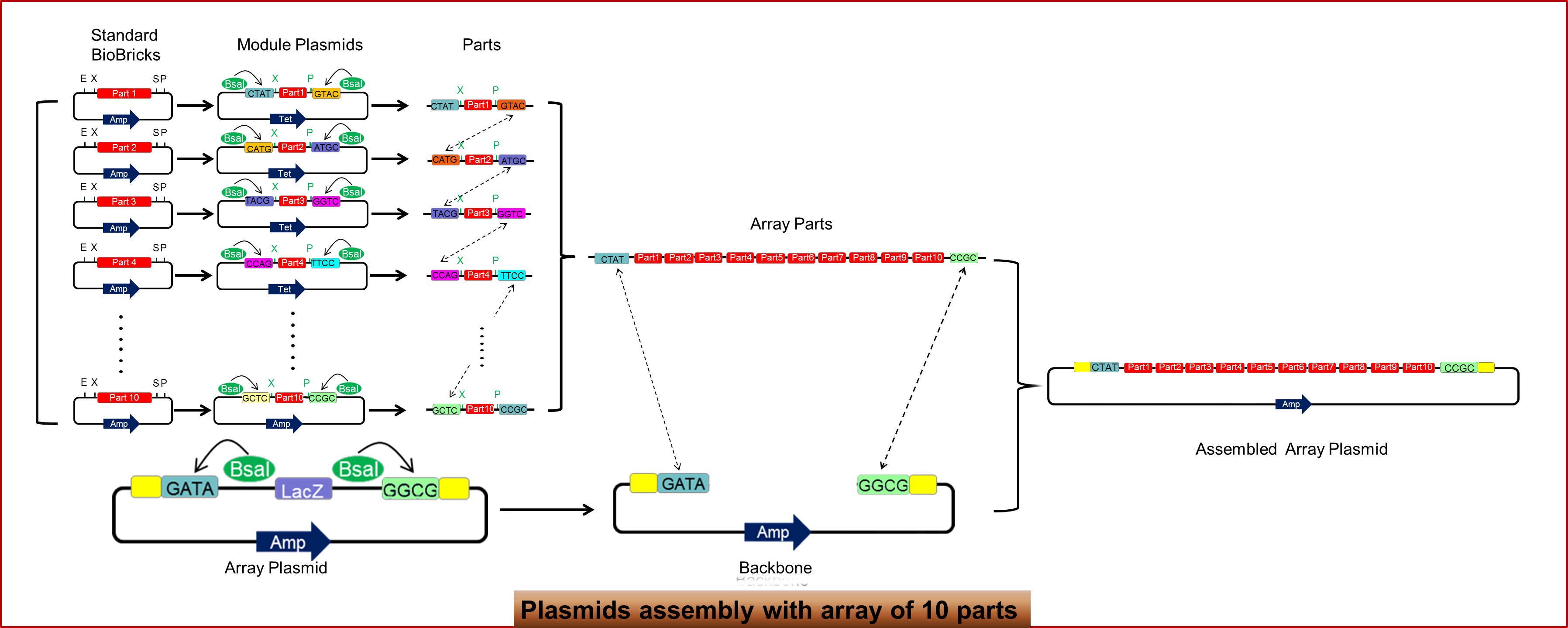
Figure 1.1 shows an overview of our design. Ten standardized biobricks are digested by Xbal and Pstl, and ligated to ten module plasmids designed beforehand. Mix these newly assembled plasmid and an array plasmid in a single reaction, and add a restriction enzyme called Bsal and ligase. By using the type IIS restriction endonuclease BsaI,we will get those ten biobricks with cohensive end nose to tail; and in the same reaction, these ten bricks will assemble in order into array plasmid. The array plasmid has lacZ flanked by Bsal recognition site, and under the effect of Bsal, it will be able to produce sticky ends complementary to the first and tenth module plasmids. The whole process will take only four days, while the traditional method will take at least six days even in the best of circumstance.

As shown in the figure 1.2, the design of module plasmids is based on a unique type IIS restriction endonuclease-- type IIS restriction endonuclease. Bsal can recognize ‘GGTCTC’ specifically, and it can cleave outside their recognition sites to create unique 4 bp overhangs. Due to this attribute of type IIS restriction endonuclease, we have designed a series of module plasmids, the sticky ends of which are complementary one by one in order. The backbones of these plasmids are identical, the only difference lies in the prefix and postfix, as shown in the figure 1.2.
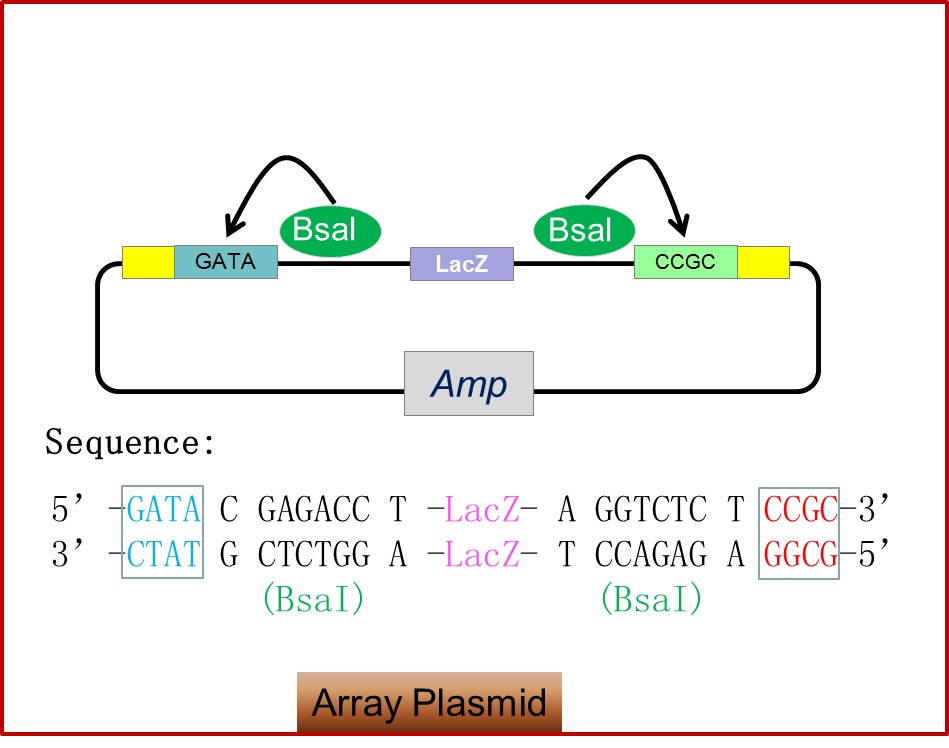
Figure 1.3 shows design of array plasmids. Array plasmids are also based on the characteristics of Bsal, but different from the module plasimids, the Bsal recognition sites are on the fragment flanking the lacZ coding sequence. After the use of Bsal, lacZ will be released with Bsal recognition sites, and the backbone can be ligated to those ten biobricks arranged in order.
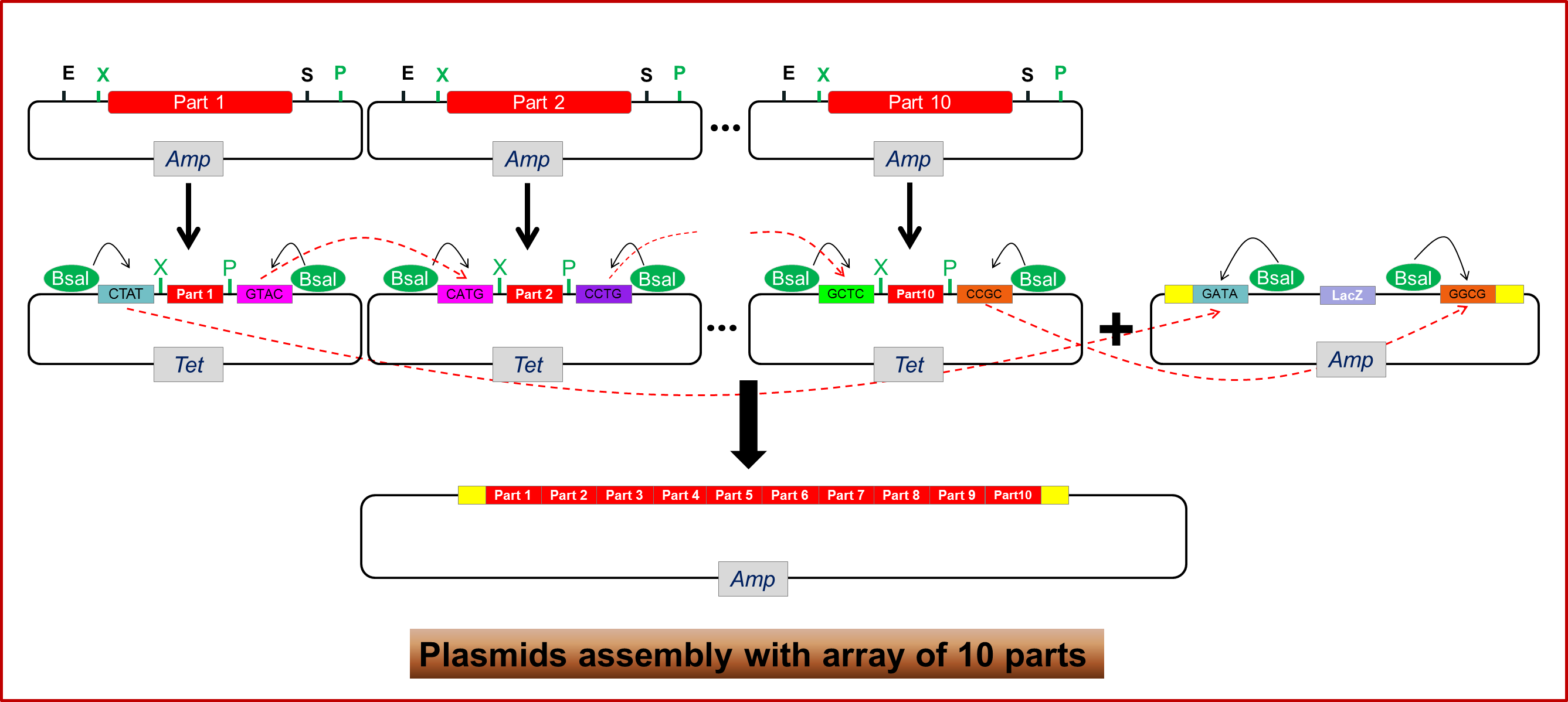
Shown as figure 1.4, we take the splicing of ten biobricks as an example. Before experiment, we should sketch the order of ten biobricks ahead. Then we put those ten biobricks on the corresponding module plasmids, digesting by Xbal and Pstl. For instance, if we want to assemble partA~partJ in order, we should put partA on module1, part B on module2, etc. Then mix those ten assembled module plasmids and array plasmid in a single reaction, and adds adequate ligase and Bsal. Incubate the reaction in a thermocycler for 10 cycles of 5 min at 37_C and 10 min at 16_C, then heated to 50_C for 5 min and then 80_C for 5 min. Those modules will be released and assembled in order into a array plasmid to create full length constructs.

Figure1.5 shows the splicing methods of assembling more than ten biobricks. Limited by the efficiency of ligase, if the number of modules surpasses ten, the error rate will increase drastically. As a result, when assembling more than ten biobricks, we will turn to another type IIS restriction endonuclease. Just as shown in the picture, we can put the recognition site on the array plasmids. When the assembly of the first step has finished, the assembled array plasmids can be considered as new module plasmids. Using Esp31 instead of Bsal to assemble those array plasmids in a way similar to which shown on figure 1.4, we will get the final products with more than 10 subparts.
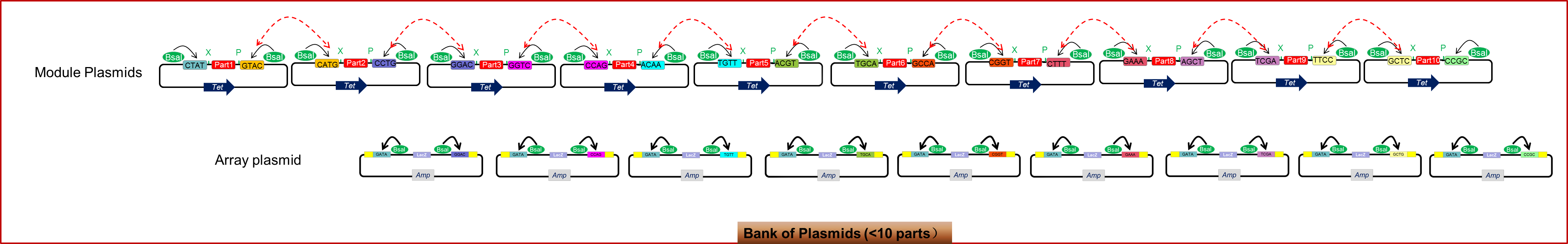
To sum up, as shown in the figure 1.6, when the system contains ten biobricks or within, we have to construct a bank of 19 plasmids, among which ten are module plasmids, and 9 are array plasmids. And this bank will make the rapid splicing of 2~10 biobricks possible. If the number of biobricks surpasses ten, we can extend the capacity of the bank based on methods shown by figure 1.5. With the aid of golden gate cloning, we can create a more standard, rapid and efficient way of assembling biobricks
Reference Adam J. Bogdanove. Daniel F. Voytas, etc. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research, Vol, 39(12), 2011.
Standard 2
As we know, biobrick is a piece of DNA that can be spliced to any other biobrick. Standard biobricks share the same backbone with four restriction endonuclease sites: EcoRI & XbaI in the prefix and SpeI & PstI in the postfix. In traditional assembly, one biobrick is cut with E & S or X & P while the other is cut with E & X or S & P correspondingly. After mixing and ligation, the two biobricks are linked together, producing a new standard biobrick with the same backbone. There is always a resistance gene in each backbone as well, which gives the bacteria resistance against chloramphenicol, kanamycin, ampicillin or tetracycline resistance gene. The smurfly delicate design offers us great convenience. However, the standard assembly left a problem: there’s no way to break down the biobricks once they are assembled. We met that problem when we got a composite device including several biobricks from Edingburgh team. We tried to get a single biobrick which we needed from it and we had nothing to do but design a specific PCR reaction which cost time and money. Therefore we hope to modify the standard backbone to make biobricks to better meet different requirements.
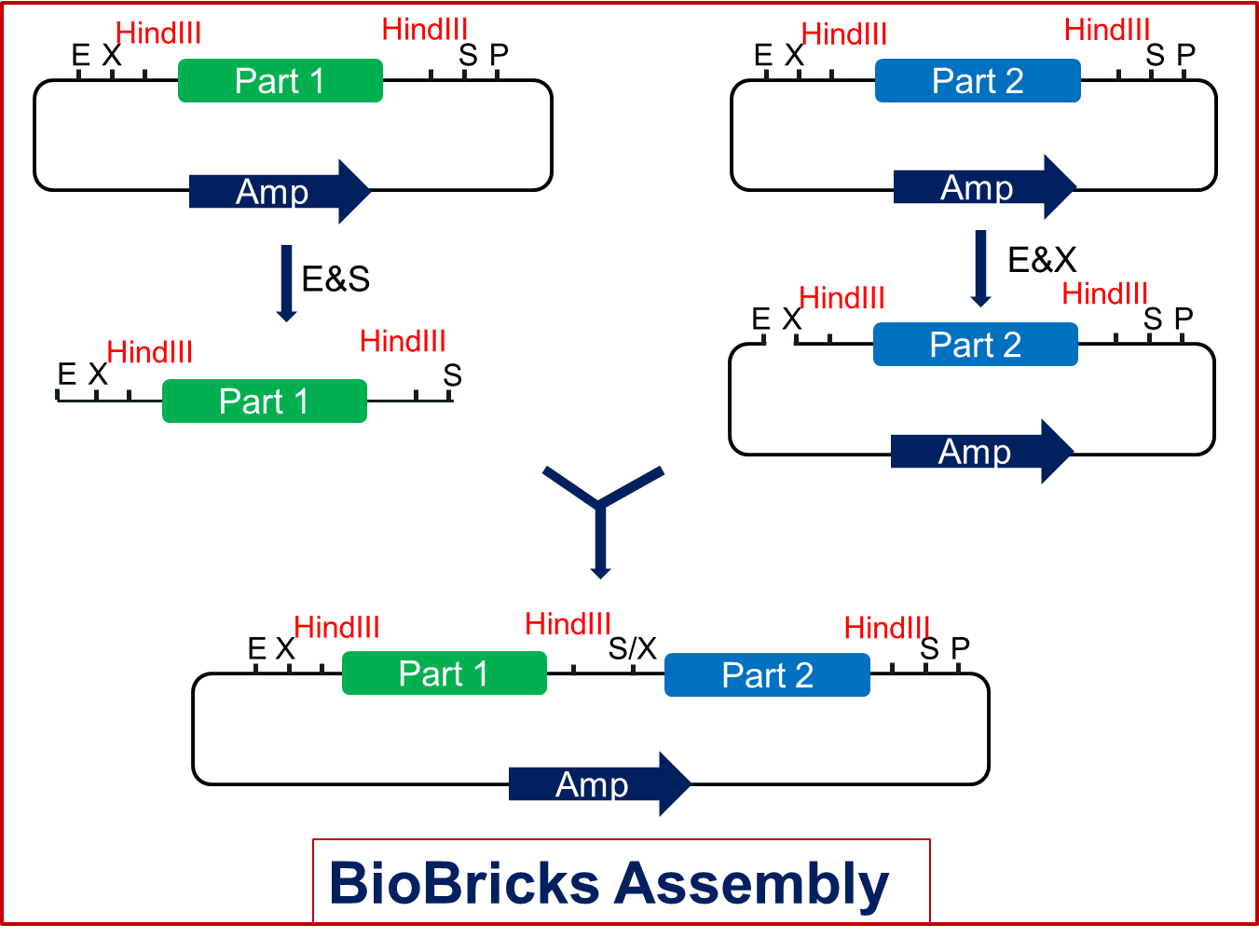
Firstly, we keep all the previous parts of the backbone so that the standard assembly still works for these new backbone plasmids (Fig2.1). Secondly, we add another restriction endonuclease site--HindIII both in the prefix and postfix of the backbones (Fig2.1). The order is shown in Fig2.1. Biobricks inserted in this kind of backbone can be assembled as the former standard. But once we produce the composite product, it will be different. Each part of the product will get two HindIII restriction sites, one upstream and the other downstream.
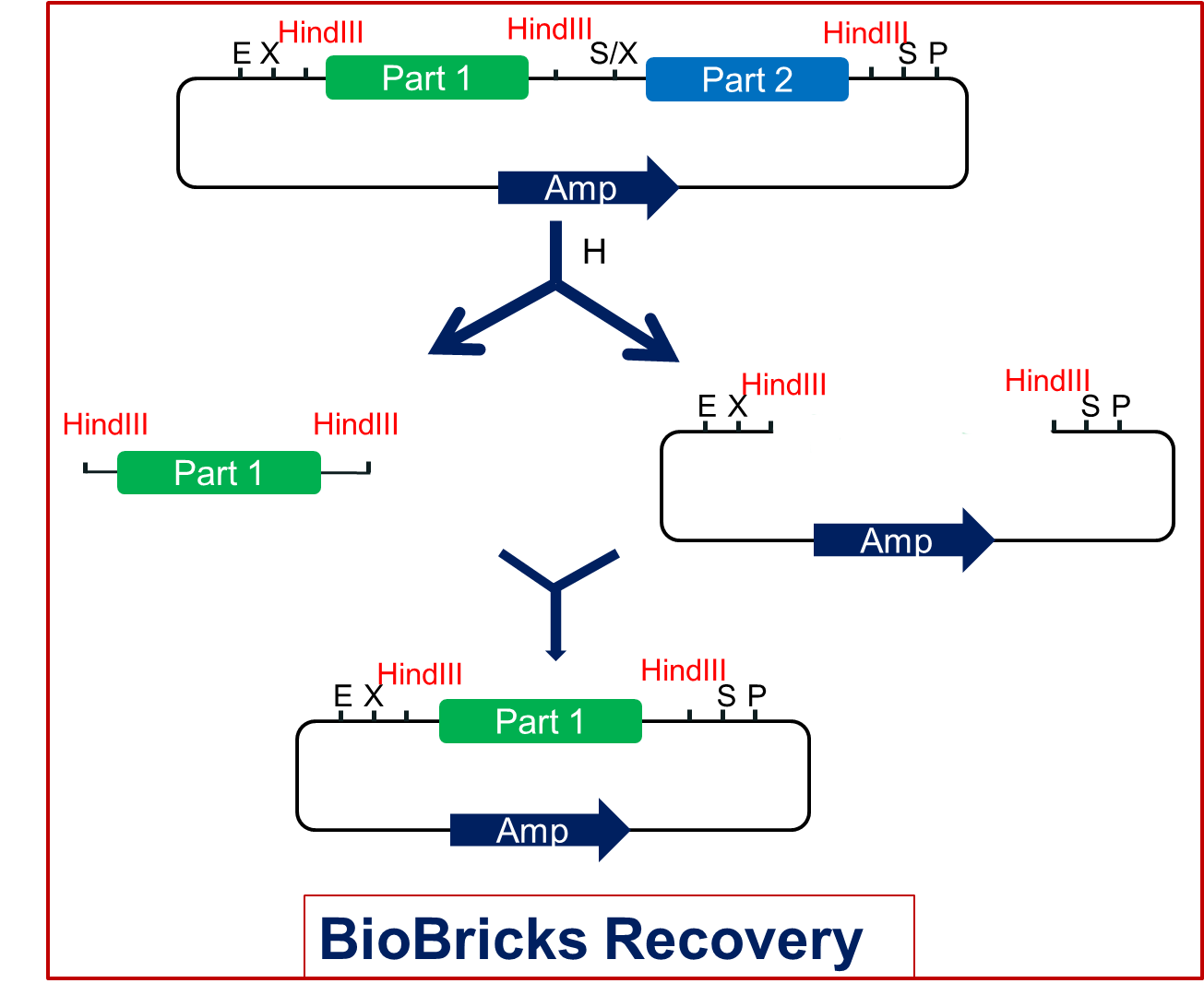
In the new assembly, we can cut this product with the HindIII restriction endonuclease and get each part as well as the vector. Then we just need to link the part with the vector. Thus we can get the single standard biobrick of each part(Fig2.2).

 "
"





