Team:WHU-China/Parts/Submitted
From 2011.igem.org
| (44 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<head> | <head> | ||
<style type="text/css"> | <style type="text/css"> | ||
| - | + | p.submit | |
{ | { | ||
| - | + | font-size:15px; | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
| - | # | + | #countdown |
{ | { | ||
| - | + | display:none; | |
| - | + | ||
} | } | ||
| - | # | + | #visit |
| + | { | ||
| + | display:none; | ||
| + | } | ||
| + | span.mw-headline | ||
| + | { | ||
| + | font-size:24px; | ||
| + | } | ||
| + | #contact | ||
| + | {position:absolute; | ||
| + | z-index:1; | ||
| + | left:0px;} | ||
| + | |||
| + | #bottom | ||
{ | { | ||
position:absolute; | position:absolute; | ||
| - | + | z-index:1; | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
} | } | ||
| - | # | + | #form |
{ | { | ||
| - | |||
| - | |||
| - | |||
position:relative; | position:relative; | ||
| + | top:180px; | ||
| + | left:20px; | ||
| + | width:960px; | ||
} | } | ||
</style> | </style> | ||
| Line 39: | Line 43: | ||
$(document).ready(function() | $(document).ready(function() | ||
{ | { | ||
| - | + | var a1=1606; | |
| + | var a2=document.body.clientHeight; | ||
| + | document.getElementById("bottom").style.top=(a2+a1).toString()+'px'; | ||
| + | document.getElementById("contact").style.top=(a2+a1+80).toString()+'px'; | ||
}); | }); | ||
</script> | </script> | ||
| Line 45: | Line 52: | ||
<body> | <body> | ||
<div id="content1"> | <div id="content1"> | ||
| - | |||
<div id="form"> | <div id="form"> | ||
| - | <img | + | <p class="submit"> These are biobricks submitted by us this year. We have already finished construction of many new biobricks and devices, but due to some problems encountered in the experiments and the lack of time, we fail to put them on the vector with chloramphenicol resistance.</p> |
| - | < | + | <h2><span class="mw-headline"> BBa_K560000: Blue Light Sensing and Reporting System </span></h2> |
| - | John T.M. Kennis and Sean Crosson. A Bacterial Pathogen Sees the Light. Science 317, 1041(2007).</p> | + | <p class="submit"> This part was designed to build the complete blue light sensing and reporting system in a single plasmid. Promoter BBa_J23117 is a constitutive promoter and not a strong one. It will keep a certain concentration of LovTAP in bacteria. When exposure to blue light, the absorption of a photon will leads to conformational rearrangements in LovTAP. The LovTAP can then bind to DNA and repress the transcription of genes following the trpL promoter. Here we use RFP as the reporter. |
| + | </p> | ||
| + | <p><br/> | ||
| + | </a><img src="/wiki/images/1/18/Whu-submitted1.png" width="509" height="279" border="0" style="margin-left:150px;" /></a> | ||
| + | </p> | ||
| + | <h2> <span class="mw-headline"> Detailed Description of LovTAP </span></h2> | ||
| + | <p class="submit"> LovTAP is a fusion protein. It consists of a LOV (light-oxygen-voltage) domain of Avena sativa phototropin1 (AsLOV2) and the trpR- DNA binding domain of the transcription factor trp repressor. | ||
| + | LOV domain is first identified as a new type of light sensor by Swartz et al in the notorious pathogen Brucella abortus. LOV domains belong to the PAS(Per-Arnt-Sim) protein domain superfamily. It has a compact protein module of about 110 amino acids. The LOV domain works through a noncovalently bound flavin cofactor when exposure to blue light. When absorbing a blue photon, the flavin cofactor will form a covalent bond with a conserved cysteine residue in the LOV domain, which results in conformational rearrangements in LovTAP(Fig 2). Flavin-binding LOV domains are fused to a range of “output” domains including bacterial diguanylate cyclases and phosphodiesterases, zincfinger, transcription factors, and histidine, kinases. | ||
| + | </p> | ||
| + | <p><br/> | ||
| + | <img src="/wiki/images/d/db/Sub2.png" width="326" height="424" border="0" style="margin-left:250px;" /> | ||
| + | </p> | ||
| + | <p class="submit"> TAP domain is trp-DNA binding domain. Here is the 3D structure of trp repressor(Fig3). Flavin-binding LOV domain changes its conformation when sensing blue light. This change impacts the affinity of the shared helix for the two domains: disrupting the contacts between the shared helix and the LOV domain and enabling the association of the shared helix with the TrpR domain, which establishes DNA-binding affinity and LovTAP can then bind DNA as an homodimer, repressing the transcription of the genes downstream of its binding sites. | ||
| + | </p> | ||
| + | <p><br/> | ||
| + | <img src="/wiki/images/0/07/Sub3.png" width="401" height="335" border="0" style="margin-left:250px;"/> | ||
| + | </p> | ||
| + | <h2><span class="mw-headline"> Resource</span></h2> | ||
| + | <p class="submit" style="font-style:Italic;"> | ||
| + | John T.M. Kennis and Sean Crosson. A Bacterial Pathogen Sees the Light. Science 317, 1041(2007). | ||
| + | </p> | ||
</div> | </div> | ||
| + | |||
| + | |||
| + | |||
| + | <div style="height:200px;"></div> | ||
| + | <div id="bottom"> | ||
| + | <image src="/wiki/images/5/58/Whu-backgroundbottom.png"/> | ||
</div> | </div> | ||
| - | <div id=" | + | <div id="contact"> |
| - | & | + | <a href="https://2011.igem.org/wiki/index.php?title=Team:WHU-China/Parts/Submitted&action=edit"><img class="whucontact" style="position:relative;left:636px;top:38px;z-index:10;" src="/wiki/images/2/2a/Whu-Home_r1_c1.png"/></a> |
| - | + | <a href="Mailto:gubrian890512@gmail.com"><img class="whucontact" style="position:relative;left:660px;top:38px;z-index:10;" src="/wiki/images/9/97/Whu-Home_r1_c3.png"/></a> | |
| - | + | <a href="https://igem.org/Main_Page"><img class="whucontact" style="position:relative;left:676px;top:38px;z-index:10;" src="/wiki/images/0/03/Whu-Home_r1_c5.png"/></a> | |
| - | </ | + | <img id="bb" src="/wiki/images/3/3c/Whu-build.jpg"/> |
</div> | </div> | ||
| + | |||
</div> | </div> | ||
</body> | </body> | ||
Latest revision as of 06:47, 27 October 2011
These are biobricks submitted by us this year. We have already finished construction of many new biobricks and devices, but due to some problems encountered in the experiments and the lack of time, we fail to put them on the vector with chloramphenicol resistance.
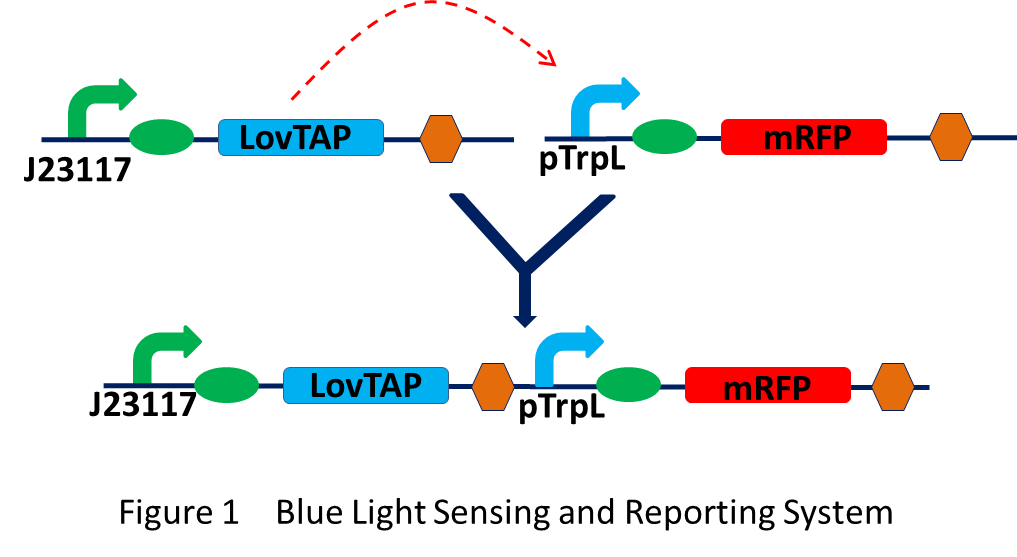
BBa_K560000: Blue Light Sensing and Reporting System
This part was designed to build the complete blue light sensing and reporting system in a single plasmid. Promoter BBa_J23117 is a constitutive promoter and not a strong one. It will keep a certain concentration of LovTAP in bacteria. When exposure to blue light, the absorption of a photon will leads to conformational rearrangements in LovTAP. The LovTAP can then bind to DNA and repress the transcription of genes following the trpL promoter. Here we use RFP as the reporter.
Detailed Description of LovTAP
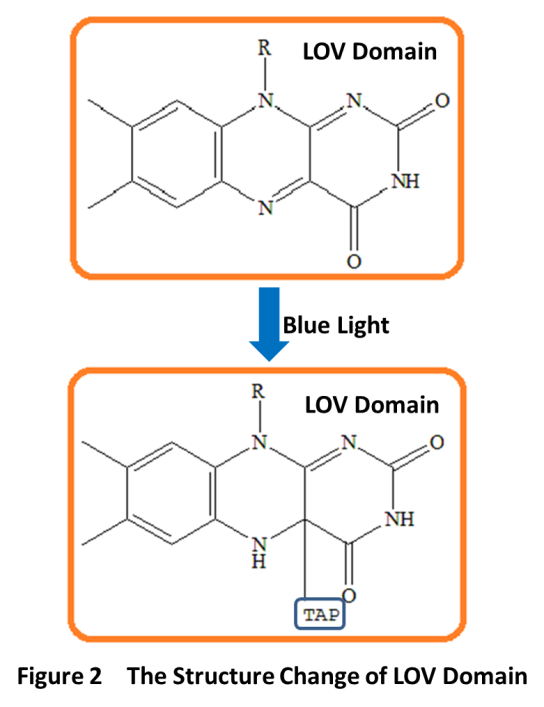
LovTAP is a fusion protein. It consists of a LOV (light-oxygen-voltage) domain of Avena sativa phototropin1 (AsLOV2) and the trpR- DNA binding domain of the transcription factor trp repressor. LOV domain is first identified as a new type of light sensor by Swartz et al in the notorious pathogen Brucella abortus. LOV domains belong to the PAS(Per-Arnt-Sim) protein domain superfamily. It has a compact protein module of about 110 amino acids. The LOV domain works through a noncovalently bound flavin cofactor when exposure to blue light. When absorbing a blue photon, the flavin cofactor will form a covalent bond with a conserved cysteine residue in the LOV domain, which results in conformational rearrangements in LovTAP(Fig 2). Flavin-binding LOV domains are fused to a range of “output” domains including bacterial diguanylate cyclases and phosphodiesterases, zincfinger, transcription factors, and histidine, kinases.
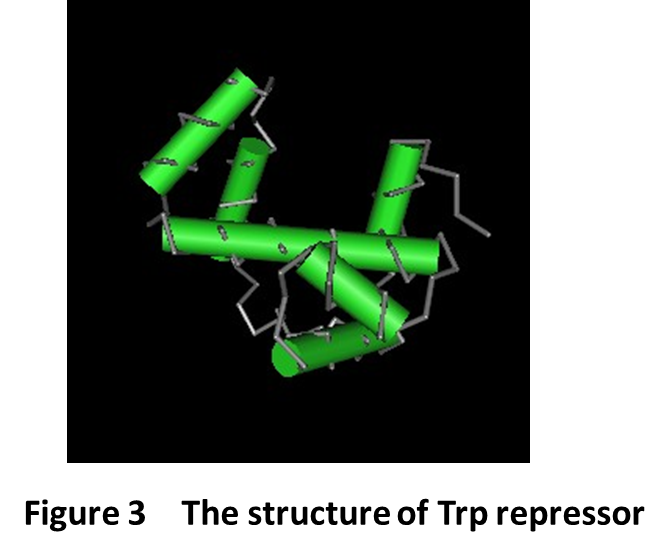
TAP domain is trp-DNA binding domain. Here is the 3D structure of trp repressor(Fig3). Flavin-binding LOV domain changes its conformation when sensing blue light. This change impacts the affinity of the shared helix for the two domains: disrupting the contacts between the shared helix and the LOV domain and enabling the association of the shared helix with the TrpR domain, which establishes DNA-binding affinity and LovTAP can then bind DNA as an homodimer, repressing the transcription of the genes downstream of its binding sites.
Resource
John T.M. Kennis and Sean Crosson. A Bacterial Pathogen Sees the Light. Science 317, 1041(2007).
 "
"





