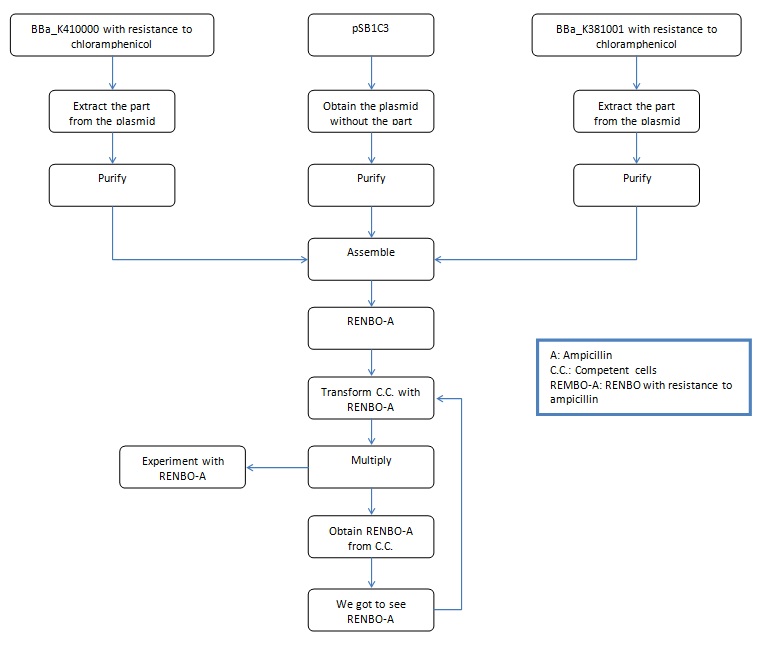
October 25, 2011
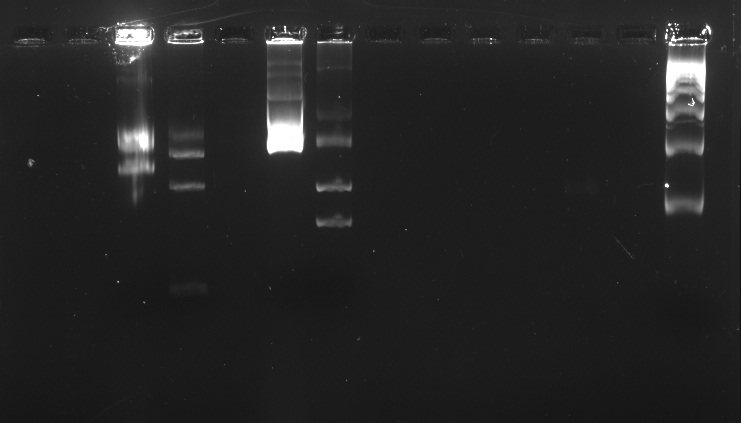
We ran an electrophoresis before we started the purification. The bands we needed were the ones with the weight mentioned.
Information necessary for the purification:
BBa_K381001: 986bp
BBa_K410000: 1517bp
pSB1C3: 2070bp
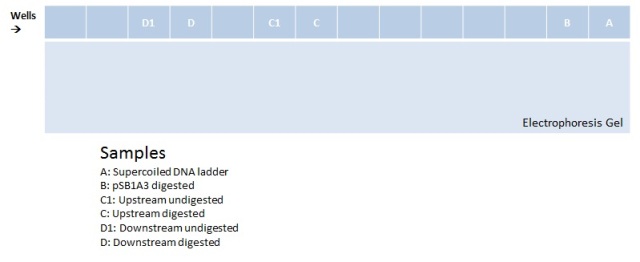
Electrophoresis with BBa_K410000, BBa_K381001, and pSB1A3 digested and undigested
Aqui va la imagen grimaldo. La celeste
Purification
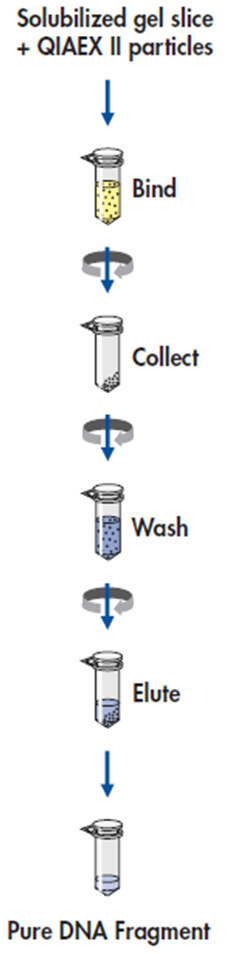
Protocol: Extraction of 40 bp to 50 kb DNA fragments from Agarose Gels
Procedure
1. DNA band was excised from the agarose gel with a clean, sharp scalpel. It was used a 2ml microfuge tube.
2. Gel slice was weighed in a colorless tube. Then added 3 volumes of Buffer QX1 to 1 volume of gel for DNA fragments 100 bp – 4 kb. For us, added 600 μl of Buffer QX1 to 200 mg of gel.
3. Re-suspended QIAEX II by vortexing for 30 s. Added 500μl QIAEX II to the sample.
4. Incubated at 50°C for 10 min to solubilize the agarose and to bind the DNA. Mixed by vortexing every 2min to keep QIAEX II in suspension. Checked that the color of the mixture is yellow.
5. Sample was centrifuged for 1min at 10000rpm and carefully removed supernatant with a pipet.
6. Pellet was washed with 500μl of Buffer QX1. Discarded supernatant. Then it was centrifuged.
7. Pellet was washed twice with 500μl of Buffer PE. Discarded supernatant. Then it was centrifuged.
8. Air-dried pellet for 10–15 min or until the pellet becomes white.
9. DNA was eluted, added 20μl of H2O and re-suspended the pellet by vortexing. This was incubated at room temperature for 5 min.
10. It was centrifuged for 1min, then pipet the supernatant into a clean tube.
PURIFICATION PROTOCOL
Bibliography:
QIAEX II® Handbook. For DNA extraction from agarose and polyacrylamide gels and for desalting and concentrating DNA from solutions. October 2008. QIAGEN Sample and Assay Technologies.
 "
"





