Team:UT-Tokyo/Project/System
From 2011.igem.org
m |
|||
| Line 17: | Line 17: | ||
The following is the gene construct we designed. | The following is the gene construct we designed. | ||
| - | + | [[File:UT_Tokyo_20-3-27-5.png]] | |
The coding sequence of aspA is inserted downstream of a substrate-responsive promoter. | The coding sequence of aspA is inserted downstream of a substrate-responsive promoter. | ||
Revision as of 10:21, 4 October 2011
System

iGEM UT-Tokyo
Project Overview
As we stated in the Background page, Our project aims at improving the working efficiency of bioremediation by increasing bacterial density near the substrate. To do this, we developed a system consisting of two types of E. coli: “guiders” detect the substrate and attract via chemoattraction "workers", performers of the remediation task, to the substrate.
The workers are then prevented from diffusing away by a cell arrest mechanism, elongating the duration of contact with the substrate.
Chemotattraction
E.coli have a native property called 'chemotaxis,' and are attracted to certain substances called 'chemoattractants.’ E.coli detect and move towards a higher concentration of the substances.
In our system we utilize this property to raise cell density. Specifically, we aim to realize substrate-responsive chemoattractant overproduction and a resultant cell assembly to the substrate.
The chemoattractant we have utilized is aspartate (L-Asp). Aspartate has previously been shown to act as a chemoattractant[1], which we confirmed by our own experiment, the swarm assay. By making E.coli discharge aspartate, an aspartate concentration gradient around these bacteria is generated. Workers are attracted, creating a higher cell density around the aspartate-secreting E.coli.
We have devised a substrate-responsive aspartate overproduction mechanism. By utilizing this mechanism, guiders overproduce and secrete aspartate in response to a particular substrate and attract other bacteria to enhance cell density around the substrate.
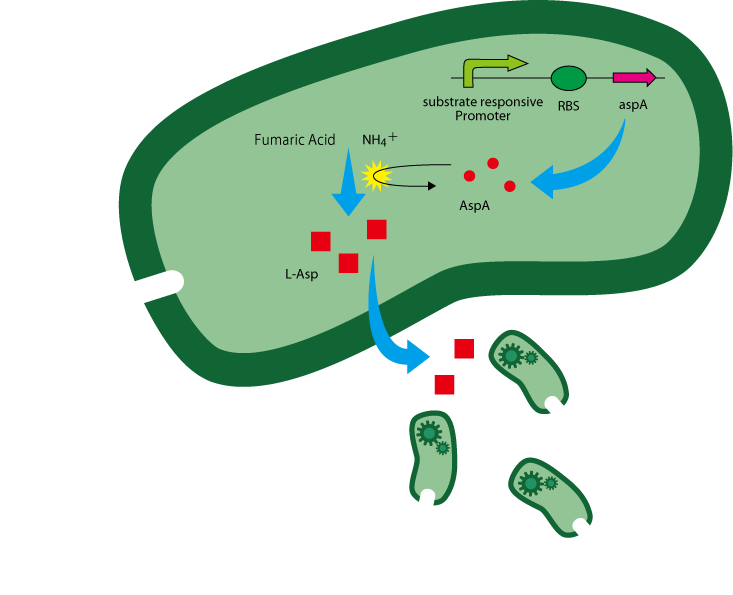
The following is the gene construct we designed.
The coding sequence of aspA is inserted downstream of a substrate-responsive promoter. The reaction of aspartate production from fumaric acid and anmonium ion is catalyzed by aspartase(AspA) encoded by the gene aspA[2]. (Figure. 1)
L-Asp is overproduced in the presence of enough AspA and the two substrates.
Note that because asp chemoattraction is native to E.coli, guiders are also attracted to the substrate. Attracted guiders also produce aspartate, giving rise to a positive feedback in which guiders are continuously added and create even higher aspartate concentrations.
Cell Arrest
As the reaction catalyzed by AspA is reversible, when the aspartate concentration around guiders becomes too high, the reaction reaches equilibrium and so additional aspartate cannot be produced. In addition, the substrates for AspA (fumaric acid and anmonium) may be limited. For these reasons, aspartate secretion by guiders does not last forever. The aspartate concentration gradient produced by guiders is gradually lost by diffusion as time elapses. Consequently, in order to maintain a high worker bacteria density, we attempted to 'arrest' the bacteria at the target location to prevent bacteria from escaping. To achive this, we have developed a mechanism that make bacteria less motile when they are at the target location by manipulating flagellar movement.
Bacterial chemotaxis and flagellar movement has been extensively investigated and the molecular basis for E.coli chemotaxis is understood. According to a previous study[3], a gene named cheZ, which codes for a protein involved in chemotactic signaling pathway, regulates cell motility. More specifically, cheZ, constitutively expressed in E.coli, phosphorylates a flagellar motor binding protein called CheY, making the cell motile. Therefore, cells are motile in the presence of cheZ and immotile in the absence of cheZ.
In order to make "worker" bacteria stay at the target site, we need to lower the cell motility in response to the substrate. Put in another way, we want cheZ to be expressed in the absense of the substrate and repressed in the presence of the substrate.
To achieve this, we utilize a cheZ knockout strain to eliminate baseline expression and devised the gene construct shown below, in which cheZ expression is repressed in a substrate-dependent manner. (Figure. 3)
When the cell does not detect the substrate, cI inhibitor is not expressed and therefore cheZ is expressed and the product rescues motility of the cheZ- strain. Once the cell detects the substrate, the substrate-responsive promoter is activated and cI inhibitor is expressed. cI inhibitor then inhibits the activity of cI promotor and thereby represses cheZ expression, which leads to substrate-induced loss of motility.
By utilizing these two mechanisms, the high worker density around the substrate is produced and maintained.
References
- [1] Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 2007;129(21):6807-6811.
- [2] Chao Y, Lo T, Luo N. Selective production of L-aspartic acid and L-phenylalanine by coupling reactions of aspartase and aminotransferase in Escherichia coli. Enzyme Microb. Technol. 2000;27(1-2):19-25.
- [3] Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 2011;9(3):153-165.
 "
"