Team:USTC-China/Drylab/riboswitch
From 2011.igem.org
(→Analysis) |
(→Aptamer Sequence Design) |
||
| (21 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:USTC-China/temp}} | {{Team:USTC-China/temp}} | ||
{{Team:USTC-China/temp2}} | {{Team:USTC-China/temp2}} | ||
| - | + | <h1>Riboswitch</h1> | |
==Abstract== | ==Abstract== | ||
<p>Riboswitches are RNA-based genetic control elements that regulate gene expression in a | <p>Riboswitches are RNA-based genetic control elements that regulate gene expression in a | ||
| Line 62: | Line 62: | ||
</p> | </p> | ||
| - | [[File:Ustc5.png| | + | [[File:Ustc5.png|400px]][[File:Ustc6.png|400px]] |
<p align="center">Figure 2B. The whole sequences except 26,27,31,32 alignment result</p> | <p align="center">Figure 2B. The whole sequences except 26,27,31,32 alignment result</p> | ||
| Line 163: | Line 163: | ||
[[File:Ustc20.png|800px]] | [[File:Ustc20.png|800px]] | ||
<p align:"center">Figure 10. The secondary structure of the adenine riboswithces</p> | <p align:"center">Figure 10. The secondary structure of the adenine riboswithces</p> | ||
| + | <h3>Results</h3> | ||
| + | <p>To find out the structure-function relationship and correlations between these aptamer sequences, we did sequences alignment which indicates several important consensus sequences and the analysis of secondary structure give us more details about the dynamic processing. | ||
| + | However, from the comparing results, we found that different aptamers have varied consensus sequences and secondary structures. Thus, the aptamer sensor sections have little or nothing in common. Therefore, we should contain the sensor section and design the standard versatile platforms in our future work.</p> | ||
| - | + | <h3>Reference</h3> | |
<p>1.Shana Topp and Justin P. Gallivan. 2006. Guiding Bacteria with Small Molecules and RNA. Jacs.</p> | <p>1.Shana Topp and Justin P. Gallivan. 2006. Guiding Bacteria with Small Molecules and RNA. Jacs.</p> | ||
<p>2.Sean A. Lynch, Shawn K. Desai, Hari Krishna Sajja, and Justin P. Gallivan. 2007. A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function. Cell.</p> | <p>2.Sean A. Lynch, Shawn K. Desai, Hari Krishna Sajja, and Justin P. Gallivan. 2007. A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function. Cell.</p> | ||
| Line 172: | Line 175: | ||
<p>6. Jean-FranÇois Lemay and Daniel A. Lafontaine. 2007. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA</p> | <p>6. Jean-FranÇois Lemay and Daniel A. Lafontaine. 2007. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA</p> | ||
<p>7. Alexander Serganov, Yu-Ren Yuan,Olga Pikovskaya,1 Anna Polonskaia, Lucy Malinina,1 Anh Tuân Phan, Claudia Hobartner, Ronald Micura, Ronald R. Breaker, and Dinshaw J. Patel. 2004.Chem. Biol.</p> | <p>7. Alexander Serganov, Yu-Ren Yuan,Olga Pikovskaya,1 Anna Polonskaia, Lucy Malinina,1 Anh Tuân Phan, Claudia Hobartner, Ronald Micura, Ronald R. Breaker, and Dinshaw J. Patel. 2004.Chem. Biol.</p> | ||
| + | ==Aptamer Sequence Design== | ||
| + | <html> | ||
| + | <p>This part presents a method that was used to design aptamer sequences that should theoretically bind the target molecule (Theophylline). The method used is based on the data analysis of collected aptamer sequences.</p> | ||
| + | <h3>Design Constrains</h3> | ||
| + | <p>Designing a aptamer sequences at low background level of the gene can be achieved by increasing or decreasing the stem and loop structures. However, there are certain constrains laid on the design.</p> | ||
| + | <h4>Constrains on the consensus sequence</h4> | ||
| + | <p>As we mentioned at the data analysis part, there is the requiments that we must contain the consensus sequences near the loop region, which is regarded as a core part in the binding process.</p> | ||
| + | <h4>Constrains on the secondary structure</h4> | ||
| + | <p>As extra constrains, we take secondary structure into account. Because, firstly, we want to keep the secondary structure as designed as close as possible to the original aptamer. Besides, we want to simplify the design process and putting the requirements into our design would greatly reduce the solution space.</p> | ||
| + | <p>The previous analysis showed us that changing the stem region would influence gene expression in the absence of the target molecule without losing their function. However, it’s unclear if it can achieve the function when we change the other part. In fact, the more we change, the bigger risk that it lose its function to bind the target molecule.</p> | ||
| + | <h3>Loop Region Design</h3> | ||
| + | <p>There are basically three original aptamer sequences loop. Since the loop region plays a core role in the binding process, we decided to keep the three sequences in the designed aptamers (Table1)(Fig 1). </p> | ||
| + | <p>TABLE 1. Three original aptamer loop region sequences </p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | Num Sequences | ||
| + | 1 AUACCAAAGC | ||
| + | 2 AUACCAGUCAGC | ||
| + | 3 AUACCAGCAUC | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2011/6/65/Lfigure1.jpg" width="250" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/9/9c/Lfigure2.jpg" width="250" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/6/6d/Lfigure3.jpg" width="250" /> | ||
| + | |||
| + | <h3>Hairpin Region Design</h3> | ||
| + | <p>Designing different hairpin sequences will form a hairpin structure with a given stability profile. Thus, we can control the leaky background gene expression and the binding process.</p> | ||
| + | <p>In fact, there are varied hairpins in original aptamers and we can iteratively add the loop to further change the free energy (Table 2).</p> | ||
| + | <p>TABLE 2. Three original aptamer hairpin sequences</p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | Sequence FreeEnergy(ΔG,kcal/mol) GC Content | ||
| + | GCAGCUGC 0.4 0.63 | ||
| + | GGCAGGACC 1.2 0.78 | ||
| + | GCAGCCGC 1.2 0.88 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <h4>Artificial aptamer hairpin Sequence</h4> | ||
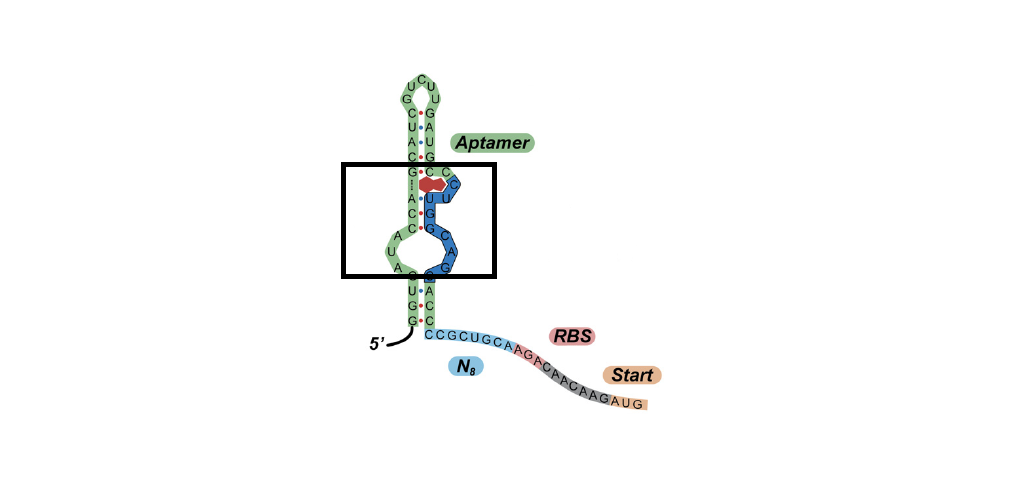
| + | <p>List all the possible hairpin sequences and see if it meets the requirements as stated above may be the most simple solution. Given the template based on the original hairpin sequences (GG***CC), there are 4 nucleotides which are free to choose. With four options per nucleotide this gives 256. For 6 nucleotides to choose it gives 4096. Indeed, only small amount of the sequences meet the requirements. Therefore, we choose some of the possible sequences and calculate the free energy.</p> | ||
| + | <p>TABLE 3. Designed hairpin sequences</p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | Sequence FreeEnergy(ΔG,kcal/mol) GC Content | ||
| + | GGUAGGACC 0.4 0.67 | ||
| + | GGAAGGACC 0.1 0.67 | ||
| + | GGCUGGACC 1.2 0.78 | ||
| + | GGCAUGACC 1.2 0.67 | ||
| + | GGCAGCCGACC 0.7 0.83 | ||
| + | GGCAGCCCGGACC -0.1 0.85 | ||
| + | GGCAGCCCCGGACC -0.1 0.86 | ||
| + | GGACCCACC 0.7 0.89 | ||
| + | GGACCCCACC 0.5 0.80 | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2011/b/bb/Lfigure4.png" width="300" /> | ||
| + | <h3>Results</h3> | ||
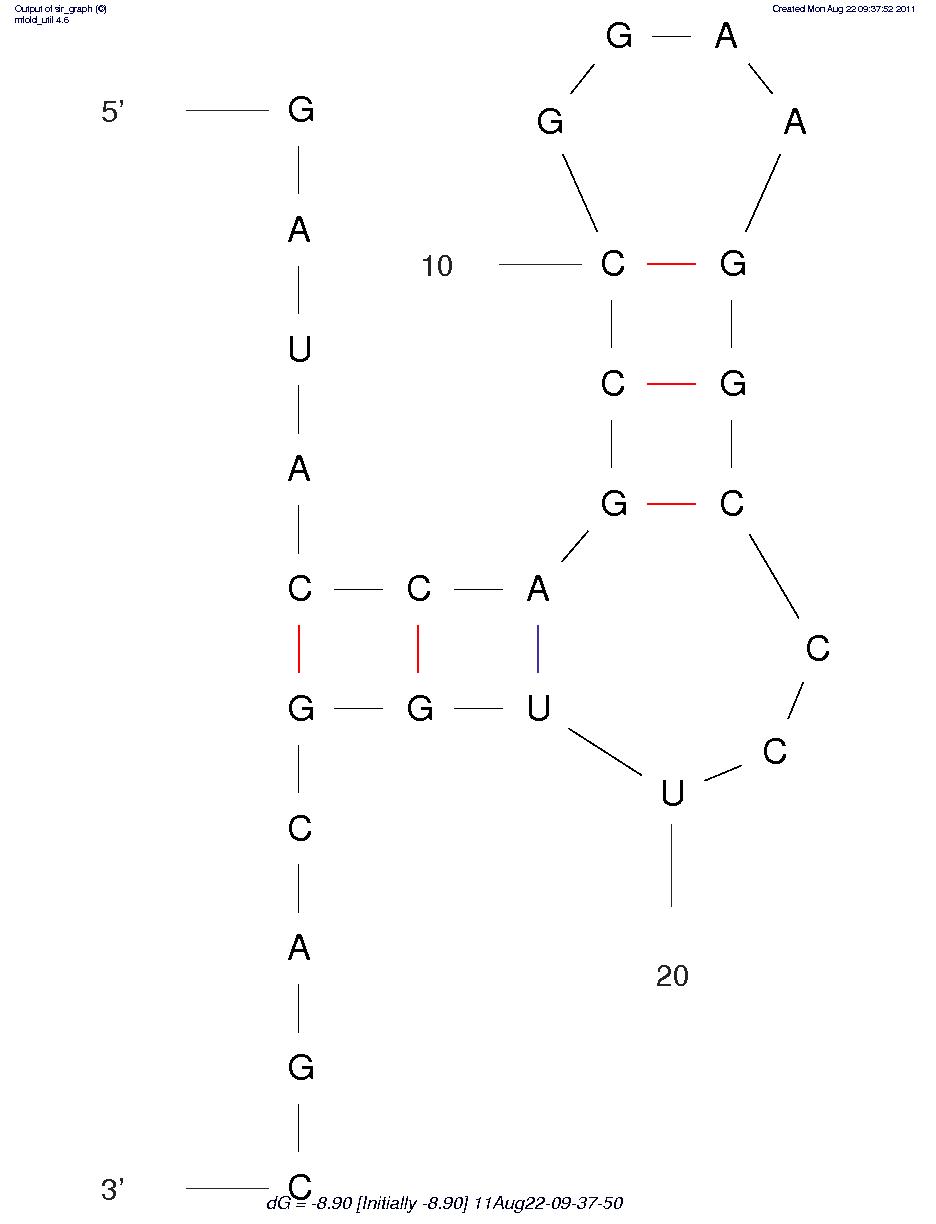
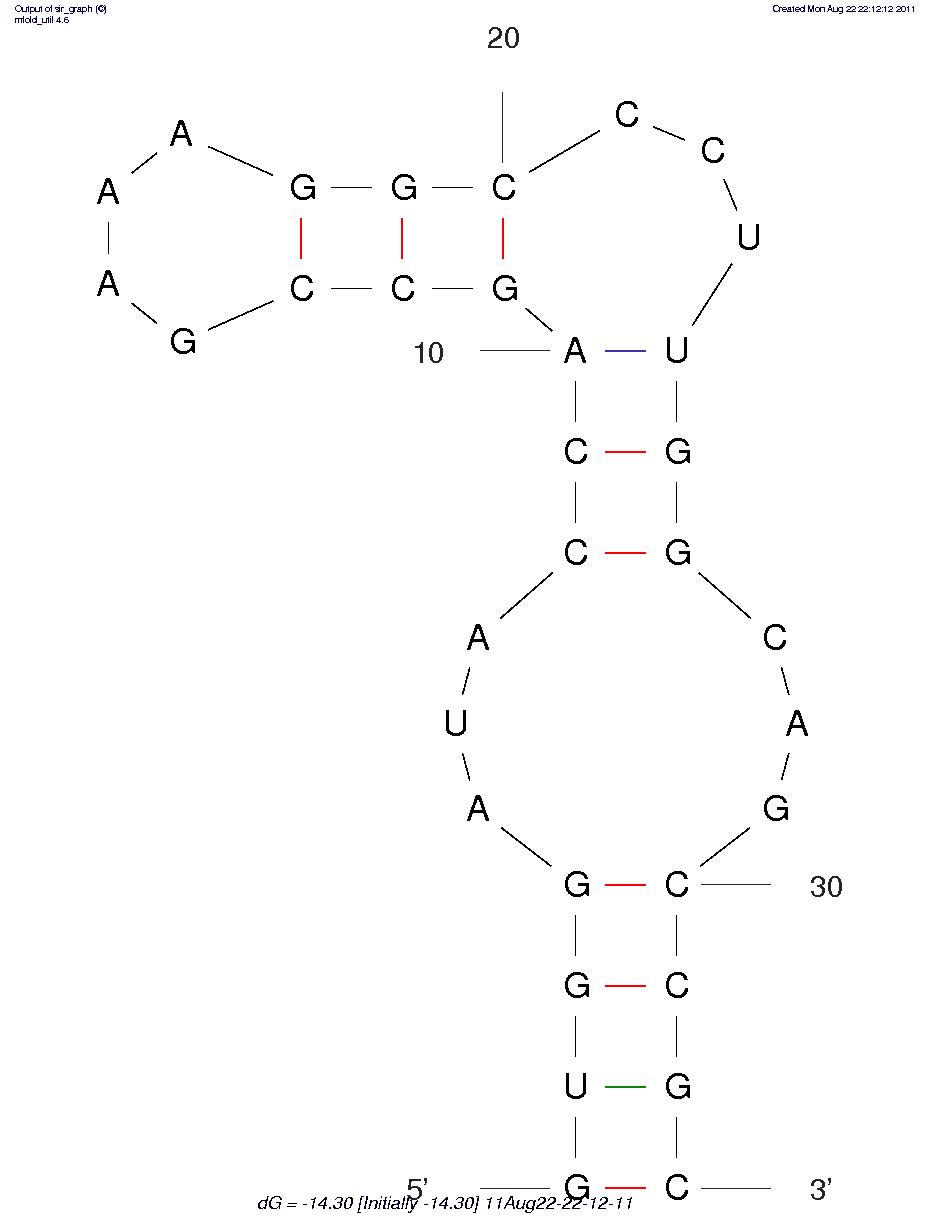
| + | <p> We combined the loop region with hairpin into the whole aptamer. And we designed varied hairpin sequences and hope that the alternative hairpin sequences with varied free energy, thus, stability profile would influence leaky background gene expression (Figure4) (Table 4).</p> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/f/fc/Lfigure5.png" width="300" /> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/1/1a/Lfigure6.png" width="300" /> | ||
| + | <p>TABLE 4. Designed sequences with varied free energy.</p> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | Sequences Num FreeEnergy(ΔG,kcal/mol) | ||
| + | Sequence 1 -8.9 | ||
| + | Sequence 2 -9.2 | ||
| + | Sequence 3 -13.7 | ||
| + | Sequence 4 -14.3 | ||
| + | Sequence 5 -14.9 | ||
| + | Sequence 6 -18.8 | ||
| + | Sequence 7 -19.2 | ||
| + | Sequence 8 -20.1 | ||
| + | Sequence 9 -23.7 | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | |||
| + | <h3>REFERENCE</h3> | ||
| + | <p>Designing RNA thermometers as part of the iGEM 2008 project. Bioinformatics Information and Communication Theory Group TU Delft</p> | ||
| + | </html> | ||
| + | <html> | ||
| + | <a href="#top"><div id="totop_box"> | ||
| + | <img class="logo" src="https://static.igem.org/mediawiki/2011/8/87/Top.png" width="50" height="50"alt="Go back to Top" style="right:auto"/> | ||
| + | </div></a> | ||
| + | </html> | ||
Latest revision as of 00:19, 29 October 2011

Contents |
Riboswitch
Abstract
Riboswitches are RNA-based genetic control elements that regulate gene expression in a ligand-dependent fashion without the need for proteins. Herein, we collect theophylline aptamers and their molecular analogues aptamers including adenine, xanthine, 3-methylxanthine, hypoxanthine and caffeine. All these sequences are available in supplemental data. We want to find out correlations among those aptamer sequences and build versatile expression platforms for required aptamer. In order to achieve these goals, we classify the aptamer sequences by analyzing their secondary structures and identify the consensus sequences by sequence alignment (BLAST engine).Finally, we reveal the riboswitches function and structure relationship and suggest design principles for creating new synthetic riboswitches and their platforms.
Analysis
Theophylline
Classify
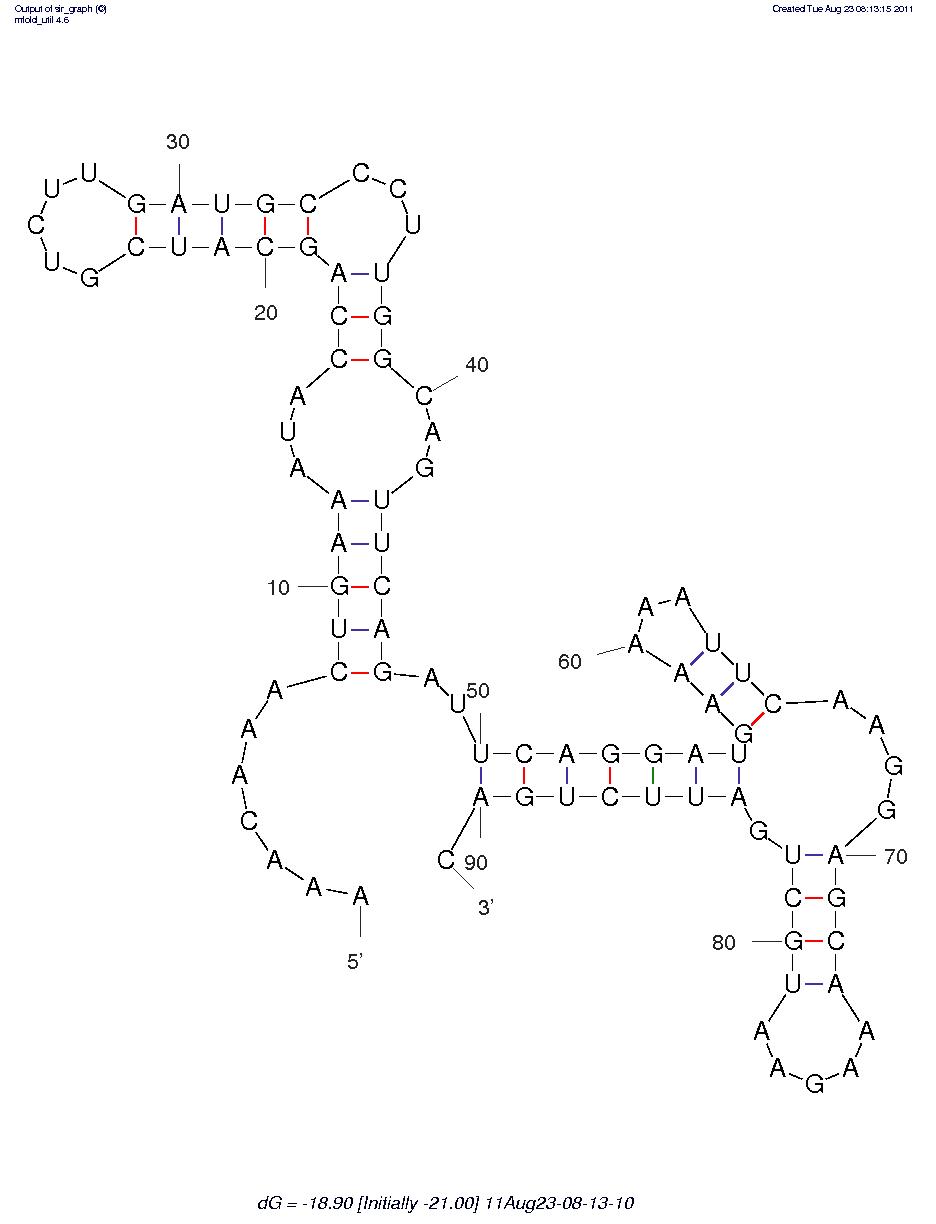
First, we predicted the secondary structure of the theophylline aptamers by mfold. Then we found that the aptamers can be classified into 3 categories by their secondary structure shapes. The first category aptamers have two loops, the second category aptamers have three loops and the last category aptamers have complex secondary structures which cannot be classified by the number of loops (Figure 1).
Figure 1A. One of the category 1 aptamer secondary structure
Figure 1B.One of the category 2 aptamer secondary structure
Figure 1C.One of the category 3 aptamer secondary structure
Sequence alignment
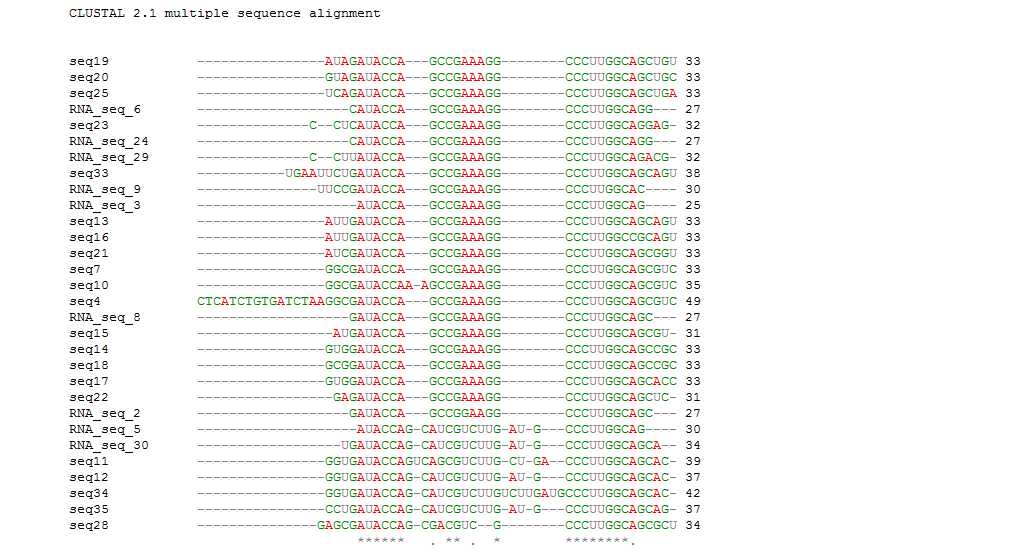
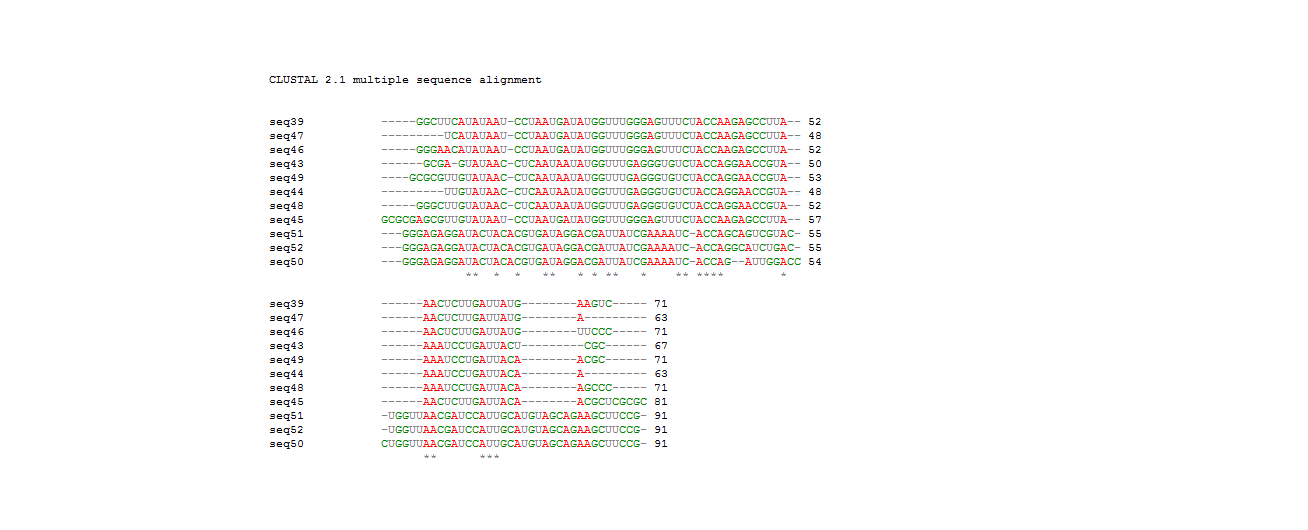
We previously got the secondary structures of the theophylline aptamers and classified those aptamer sequences into 3 categories. To find out the consensus sequences, we did the sequence alignment in each categories by Clastalw2 and the alignment indicated that the conserved sequences have varied length in different groups (Table 1)(Figure 2B). The alignment results are available in the supplemental data.
TABLE 1. Consensus sequences finds in sequences alignment
| Consensus sequence1 | Consensus sequence1 | |
| Consensus sequence1 | AUACCAGC | GCCCUUGGCA |
| Category2 | AUACCAG | GCCCUUGGCA |
| Category3 | CG | CUUGGCA |
| Whole sequences | C | GG |
| Whole sequences(except seq26, seq27,seq31,seq32) | AUACCA | CCCUUGGC |
Then we compared the whole aptamer sequences and their conserved sequences. From this alignment, we identified that the conserved sequences have the same location in the whole aptamers and the aptamers do have the same consensus sequences except several complex long aptamers. Notably, the previous work by Gallivan has reported the same motif sequences (Figure 2).
Figure 2A.The two motifs have been reported by Gallivan.
A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function DOI 10.1016/j.chembiol.2006.12.008
Figure 2B. The whole sequences except 26,27,31,32 alignment result
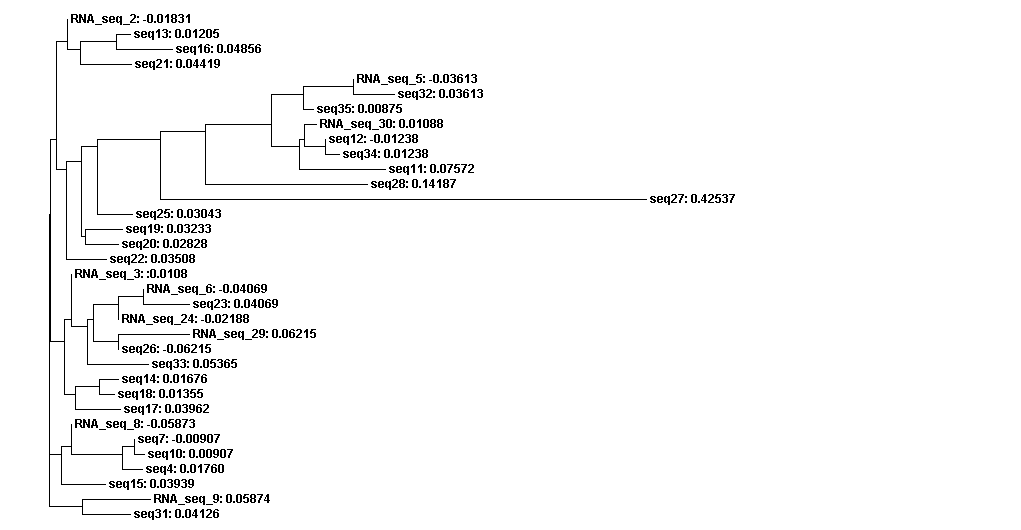
Phylogenetic analysis
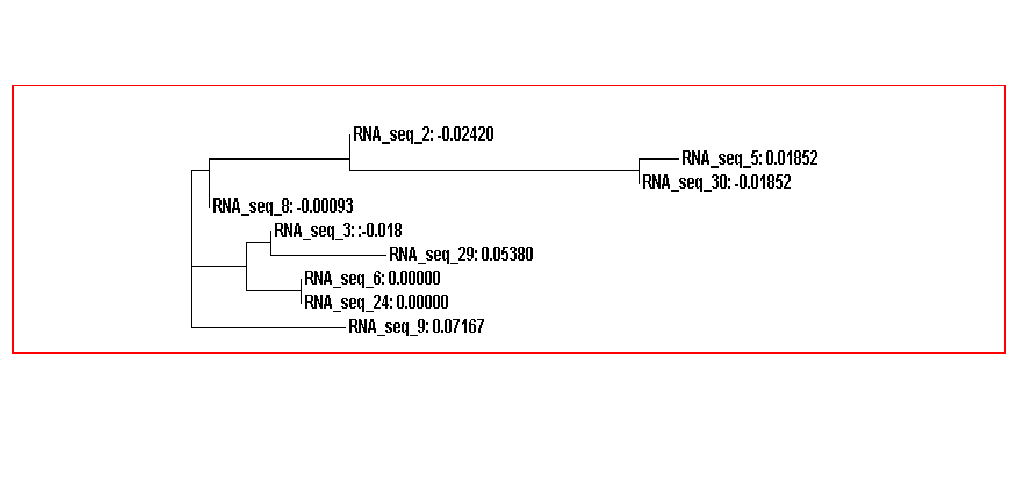
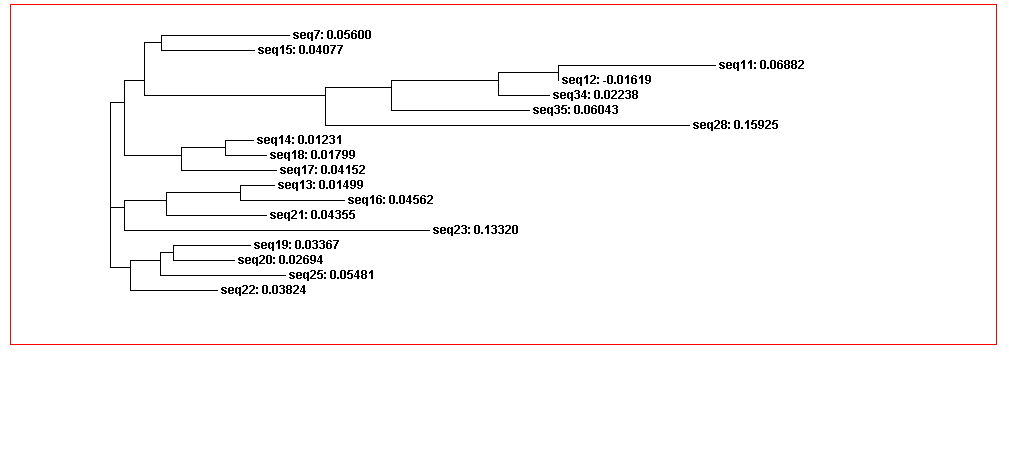
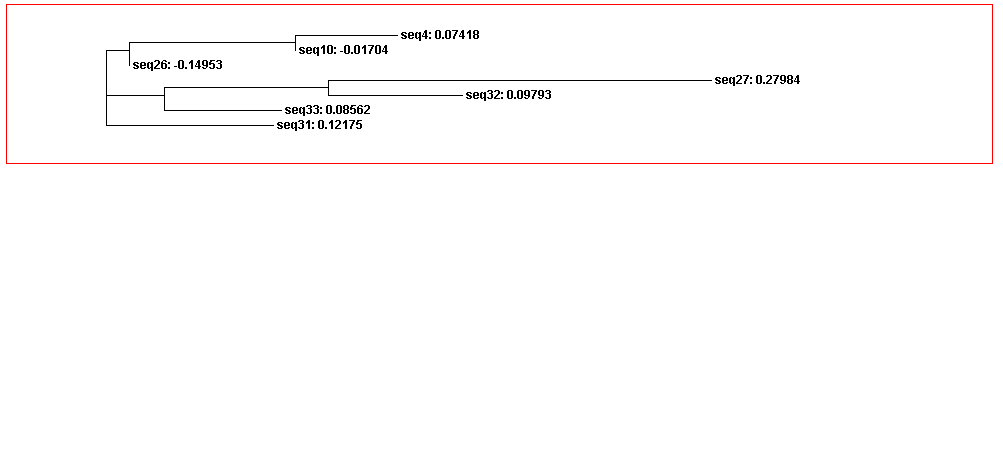
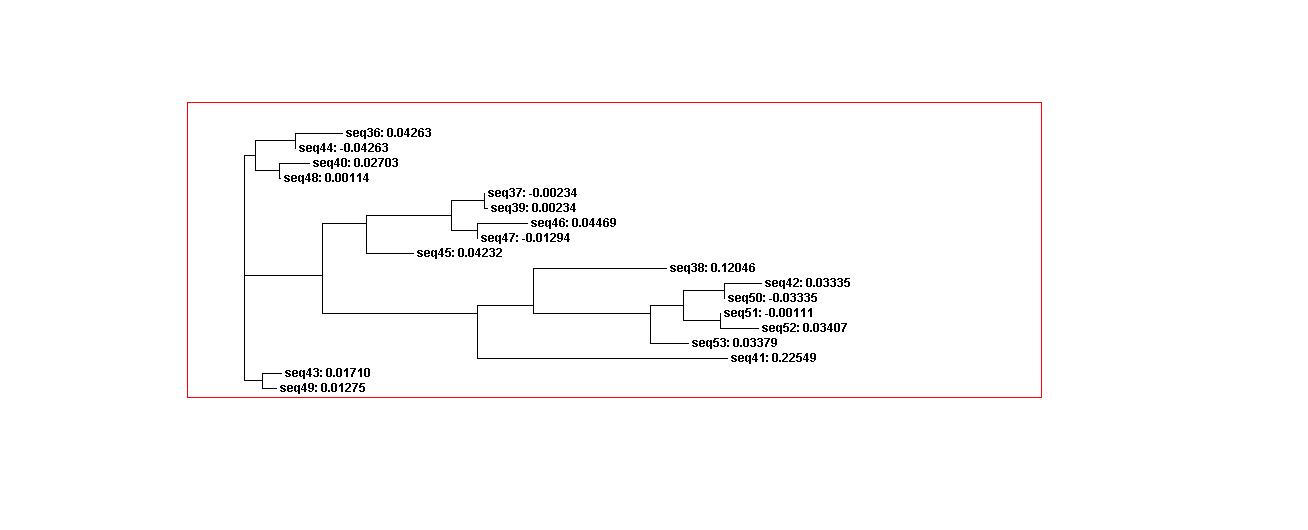
The guide tree is a branching diagram(tree), assumed to be an estimated of a phylogeny, branch lengths are proportional to the amount of inferred evolutionary change (Figure 3).
From the guide tree diagram 1 and 2, we found that the sequences are similar to each other except sequence 5 and sequence 30. Actually, the sequences alignment also indicates the sequences have relatively long consensus sequences. In guide tree diagram 3, there are less sequences than diagram 1 and 2. And the consensus sequences are quite short since the comparatively long sequences in diagram 3 are different, especially sequence 27.
From the whole sequence guide tree diagram, we found that sequences in category 1 are mixed with sequences in category 2. Thus, the classification by the sequences secondary structure may not restrict in the phylogenetic analysis perspective. The sequence 27 which is different from the other sequences in diagram 3 shows evolutionary change in the whole sequences diagram.
Figure 3A. Guide tree 1
Figure 3B. Guide tree 2
Figure 3C. Guide tree 3
Figure 3D. Whole sequences guide tree
Secondary structure and free energy
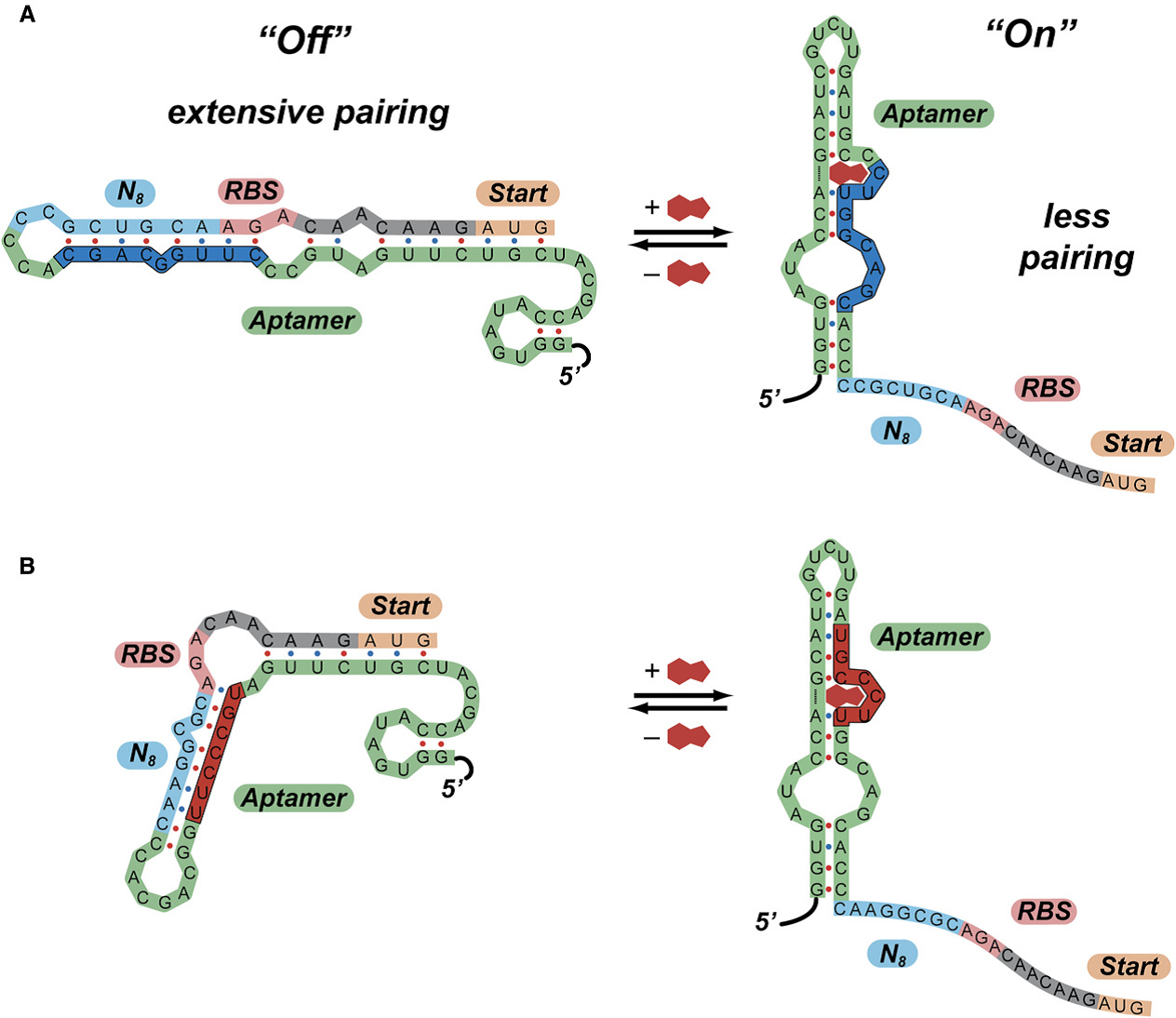
The previous work by Gallivan has shown that the aptamers usually have two conformations with different free energy and the presence of the theophylline would change the equilibrium state which the aptamers secondary structures would shift from ‘’off’’ state to ‘’on’’ state (Figure 4). The possible switching mechanism in which extensive paring in the region near the ribosome binding site prevents translation of downstream genes have been demonstrated by de Smit and van Duin.
Therefore we reconsider our previous work and the result suggests that only relative long aptamer sequence would fold to different conformation. Besides, the longer the sequences are the lower free energy the aptamer would have. And the most aptamer sequences we have collected are only the target sense section which can bind the target molecule but cannot fold into different conformations. That is, we should design the action section to accomplish the aptamer’s function.
Combing to the sequence alignment, we find that the consensus sequences locates in the region which would interact with the target molecule. As we expected, the results suggests the ligand-binding conform to the structure –function relationship.
Figure 4. Predicted Mechanisms of Action of Synthetic Riboswitches
A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function DOI 10.1016/j.chembiol.2006.12.008
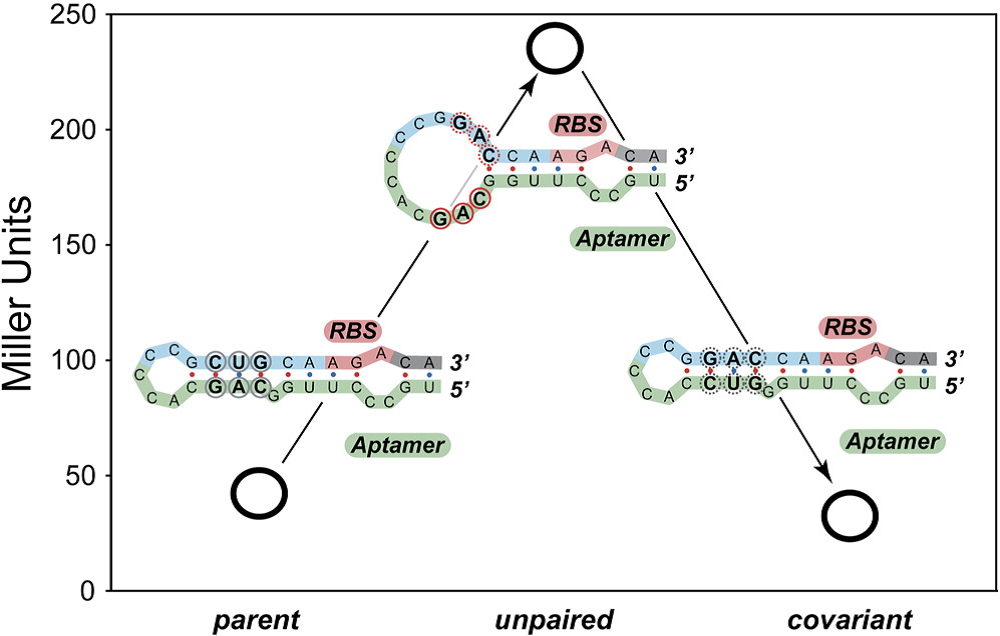
The covariance experiment by Gallivan indicates that base-pairing between the aptamer and a region near the ribosome binding would decrease the upstream gene expression which would be used to improve the aptamers’ performance (Figure 5). Notably, we use the mfold to predict the secondary structure of the aptamer which has been reported by Gallivan but cannot get the expected two varied conformation.
Figure 5. Results from Covariance Experiments
A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function DOI 10.1016/j.chembiol.2006.12.008
Adenine
Classify
As the same to the theophylline, we use mfold to predict the secondary structure of the adenine. From the results, we classified the aptamer sequences into 2 categories by their secondary structure. The sequences in the first category have the same secondary structures which have been usually reported and studied as the adenine apatamers. The sequences in the second category have varied conformation and owing to their relatively long sequence, they can fold to complex conformations.
Figure 6A.One of the category1 aptamer secondary structure
Figure 6B.One of the category2 aptamer secondary structure
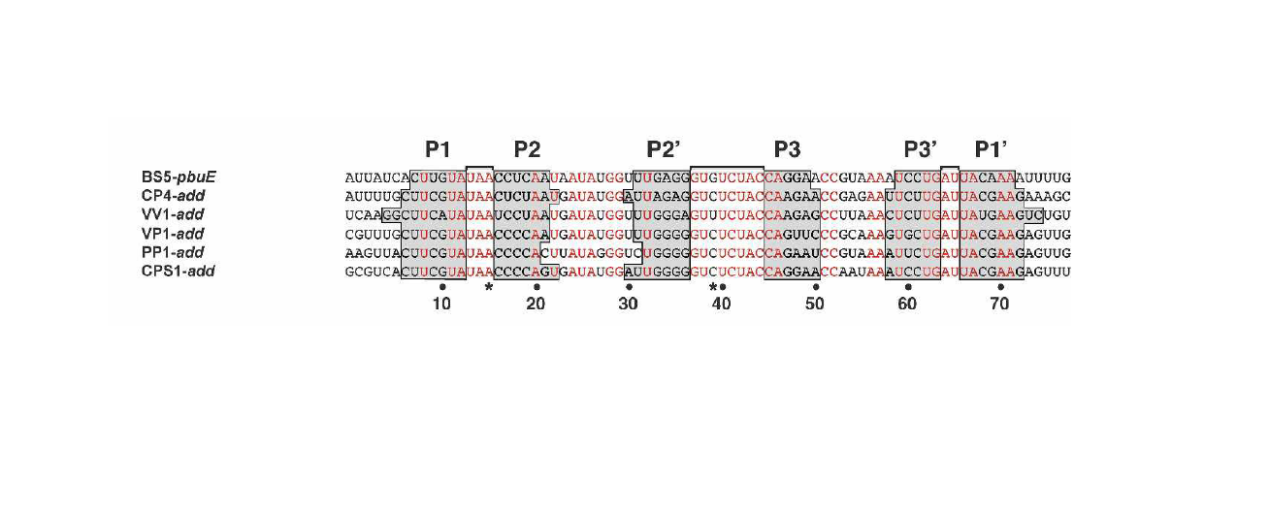
Sequence alignment
To locate the consensus sequences and probe correlation between adenine apatmer sequences and theophylline aptamer sequences, we did sequence alignment in each categories and whole sequences (Table 2) (Figure7) . From the alignment result, we found that the sequences in category 2 are quite different and have relative short consensus sequences. We thought the data which have been collected are not well processed.
We compared the adenine aptamer sequences alignment results with theophylline, we found that there are more consensus sequences locating in different region and the entire adenine aptamer sequence show complex structure. Thus, the comparison indicates that adenine aptamer sensor domain and theophylline have less or nothing in common.
In addition, the adenine alignment has shown the same consensus sequences as Daniel A. Lafontaine reported(Figure 8) Who has pointed out the importance position in adenine aptamer sequences.
TABLE 2. Consensus sequences finds in sequences alignment Consensus sequence1 Consensus sequence2 Consensus sequence3
Category1 U| Consensus sequence1 | Consensus sequence2 | Consensus sequence3 | |
| Category1 | UAUAA | GU | UGAUUA |
| Category2 | U | U | UUA |
| Whole sequences | A | \ | UU |
| Whole sequences except 36,37,38,41,42,53 | A | UU | AUU |
The consensus sequences 1,2,3 locate around 15,39,65 reported by Daniel A. Lafontaine
Figure 7. The whole sequences except 36,37,38,41,42,53 alignment result
Figure 8A. The sequence alignment result by Daniel A. Lafontaine
Core requirements of the adenine riboswitch aptamer for ligand binding RNA 2007 13: 339-350 originally published online January 2, 2007
Figure 8B. The important position in adenine aptamer sequence
Core requirements of the adenine riboswitch aptamer for ligand binding RNA 2007 13: 339-350 originally published online January 2, 2007
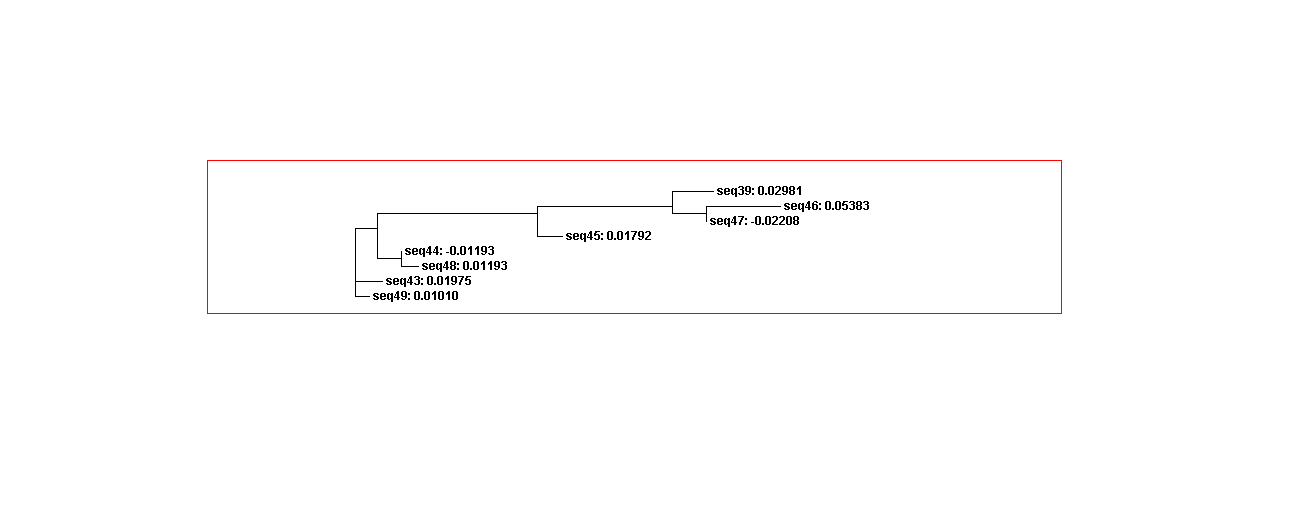
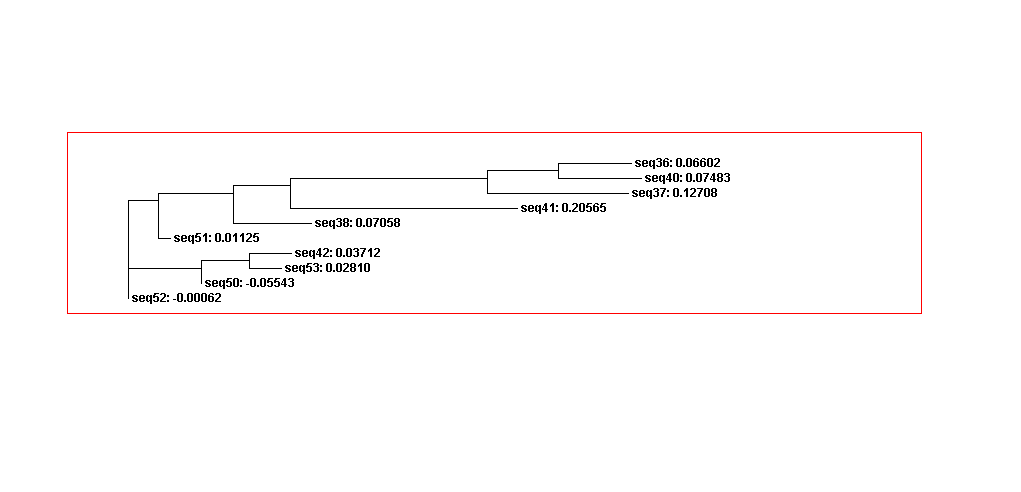
Phylogenetic analysis
Using Clustalw2, we could describe adenine aptamer sequences evolution change. Thus, sequences in category 1 more similar than category 2 since the sequences in category 1 have the same secondary structures. However, the sequences in category 1 and 2 mixes in the whole sequences guide trees. Since the adenine aptamers have completely different secondary structures from theophylline, we didn’t compare adenine aptamer with theophylline.
Figure 9A. Guide tree 1. (Adenine)
Figure 9B. Guide tree 2(Adenine)
Figure 9C. Guide tree whole sequences(Adenine)
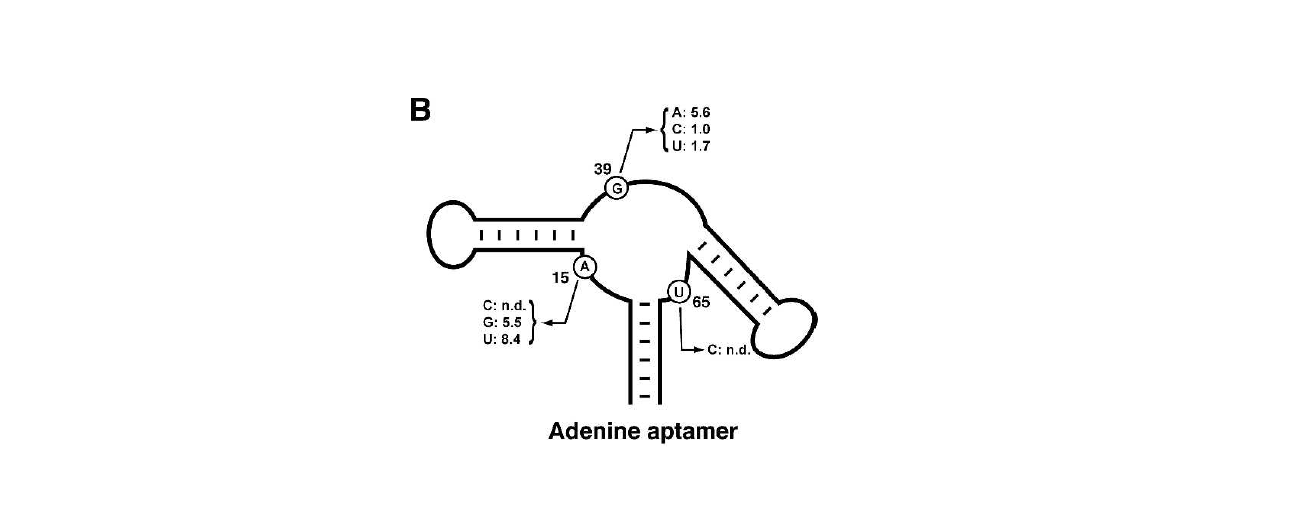
Secondary structure and free energy
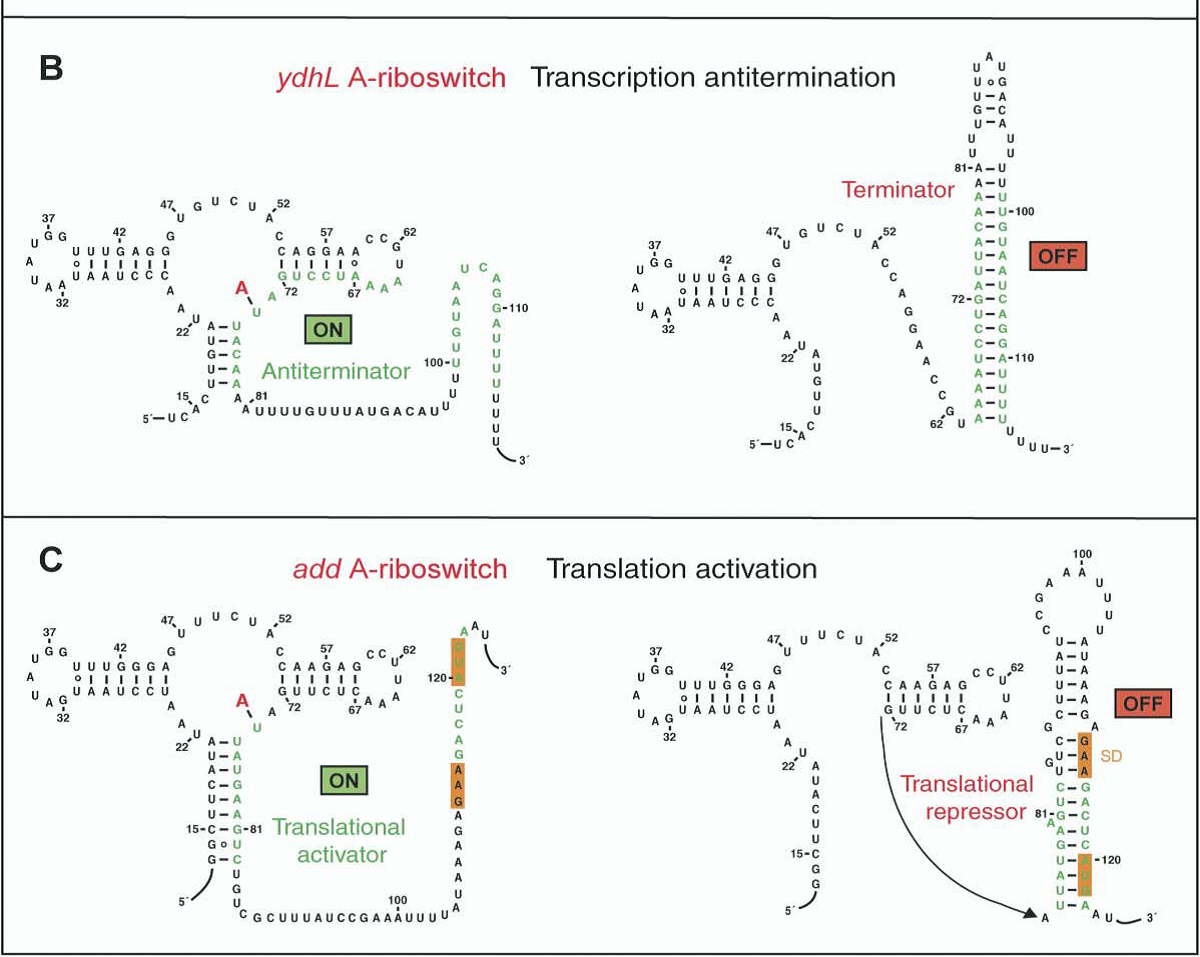
Two representatives of the adenine aptamers have been reported (Figure10). One is ydhL A-riboswitch and the other is add A-riboswitch. Both of them have the same secondary structure which have been demonstrated by our previous prediction (Figure 6A).
A remarkable feature of the ligand bound scaffolds is the formation of a junctional core involving five stacked triples (Ronald R. Breaker,3 and Dinshaw J. Patel1et al.2004). The crystal structures show that the bound ligand is directly involved in a Watson-Crick base pair at position 65 making this residue particularly important. Our sequence alignment also indicates that position 65 is consensus sequence.
Notably, the structure-function relationship in adenine aptamer would be more precisely described by their crystal structure nut not the secondary structure. In fact, we cannot predict the two conformation of the adenine aptamer by mfold.
Figure 10. The secondary structure of the adenine riboswithces
Results
To find out the structure-function relationship and correlations between these aptamer sequences, we did sequences alignment which indicates several important consensus sequences and the analysis of secondary structure give us more details about the dynamic processing. However, from the comparing results, we found that different aptamers have varied consensus sequences and secondary structures. Thus, the aptamer sensor sections have little or nothing in common. Therefore, we should contain the sensor section and design the standard versatile platforms in our future work.
Reference
1.Shana Topp and Justin P. Gallivan. 2006. Guiding Bacteria with Small Molecules and RNA. Jacs.
2.Sean A. Lynch, Shawn K. Desai, Hari Krishna Sajja, and Justin P. Gallivan. 2007. A High-Throughput Screen for Synthetic Riboswitches Reveals Mechanistic Insights into Their Function. Cell.
3. de Smit, M.H., and van Duin, J. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. USA 87, 7668–7672.
4. de Smit, M.H., and van Duin, J. 1994. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 244, 144–150.
5.de Smit, M.H., and van Duin, J. (1994). Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J. Mol. Biol. 235, 173–184.
6. Jean-FranÇois Lemay and Daniel A. Lafontaine. 2007. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA
7. Alexander Serganov, Yu-Ren Yuan,Olga Pikovskaya,1 Anna Polonskaia, Lucy Malinina,1 Anh Tuân Phan, Claudia Hobartner, Ronald Micura, Ronald R. Breaker, and Dinshaw J. Patel. 2004.Chem. Biol.
Aptamer Sequence Design
This part presents a method that was used to design aptamer sequences that should theoretically bind the target molecule (Theophylline). The method used is based on the data analysis of collected aptamer sequences.
Design Constrains
Designing a aptamer sequences at low background level of the gene can be achieved by increasing or decreasing the stem and loop structures. However, there are certain constrains laid on the design.
Constrains on the consensus sequence
As we mentioned at the data analysis part, there is the requiments that we must contain the consensus sequences near the loop region, which is regarded as a core part in the binding process.
Constrains on the secondary structure
As extra constrains, we take secondary structure into account. Because, firstly, we want to keep the secondary structure as designed as close as possible to the original aptamer. Besides, we want to simplify the design process and putting the requirements into our design would greatly reduce the solution space.
The previous analysis showed us that changing the stem region would influence gene expression in the absence of the target molecule without losing their function. However, it’s unclear if it can achieve the function when we change the other part. In fact, the more we change, the bigger risk that it lose its function to bind the target molecule.
Loop Region Design
There are basically three original aptamer sequences loop. Since the loop region plays a core role in the binding process, we decided to keep the three sequences in the designed aptamers (Table1)(Fig 1).
TABLE 1. Three original aptamer loop region sequences
Num Sequences 1 AUACCAAAGC 2 AUACCAGUCAGC 3 AUACCAGCAUC



Hairpin Region Design
Designing different hairpin sequences will form a hairpin structure with a given stability profile. Thus, we can control the leaky background gene expression and the binding process.
In fact, there are varied hairpins in original aptamers and we can iteratively add the loop to further change the free energy (Table 2).
TABLE 2. Three original aptamer hairpin sequences
Sequence FreeEnergy(ΔG,kcal/mol) GC Content GCAGCUGC 0.4 0.63 GGCAGGACC 1.2 0.78 GCAGCCGC 1.2 0.88
Artificial aptamer hairpin Sequence
List all the possible hairpin sequences and see if it meets the requirements as stated above may be the most simple solution. Given the template based on the original hairpin sequences (GG***CC), there are 4 nucleotides which are free to choose. With four options per nucleotide this gives 256. For 6 nucleotides to choose it gives 4096. Indeed, only small amount of the sequences meet the requirements. Therefore, we choose some of the possible sequences and calculate the free energy.
TABLE 3. Designed hairpin sequences
Sequence FreeEnergy(ΔG,kcal/mol) GC Content GGUAGGACC 0.4 0.67 GGAAGGACC 0.1 0.67 GGCUGGACC 1.2 0.78 GGCAUGACC 1.2 0.67 GGCAGCCGACC 0.7 0.83 GGCAGCCCGGACC -0.1 0.85 GGCAGCCCCGGACC -0.1 0.86 GGACCCACC 0.7 0.89 GGACCCCACC 0.5 0.80

Results
We combined the loop region with hairpin into the whole aptamer. And we designed varied hairpin sequences and hope that the alternative hairpin sequences with varied free energy, thus, stability profile would influence leaky background gene expression (Figure4) (Table 4).


TABLE 4. Designed sequences with varied free energy.
Sequences Num FreeEnergy(ΔG,kcal/mol) Sequence 1 -8.9 Sequence 2 -9.2 Sequence 3 -13.7 Sequence 4 -14.3 Sequence 5 -14.9 Sequence 6 -18.8 Sequence 7 -19.2 Sequence 8 -20.1 Sequence 9 -23.7
REFERENCE
Designing RNA thermometers as part of the iGEM 2008 project. Bioinformatics Information and Communication Theory Group TU Delft

 "
"