Team:UNAM-Genomics Mexico/Modeling/FBA
From 2011.igem.org
Background
Our synthetic pathway produces Hydrogen. Our chassis fixes Nitrogen, which is influenced by Hydrogen availability. Therefore, we were keenly interested in finding out how our synthetic pathway would interact with the host metabolism. After some exploratory consultation, we determined Flux Balance Analysis was an effective tool for this.
Contents |
Introduction
Flux Balance Analysis (FBA) can serve to explore the fluxes of a given metabolic reconstruction. In this case, we wanted to determine the level and extent of interaction of our added pathway with the core metabolism. Since our chassis, Rhizobium etli, has two "flavors" (free living organism & plant symbiont), we were curious as to weather our transgenic system would remain functional under symbiont form. Moreover, since a key aspect of the project was Nitrogen fixation, we wanted to ensure said pathway was as functional as it could be. And on top of that, we have to remind ourselves that our chassis is a symbiont to the photosynthetic plant Phaseolus vulgaris (the common bean), which maintains the niche in exchange for nitrous compounds.
In other wording, our chassis looks like this:
Furthermore, key researchers at our University warned us against the project due to the widespread presence of endogenous hydrogenases (the word "impossible" was often used, which only got us rebelliously stuck on proving them wrong). These native enzymes play the opposite role of the activity we were interested in, they consume molecular hydrogen instead of producing it. Apparently, since Nitrogen fixation produces molecular hydrogen, some Rhizobial species developed capture hydrogenases to recycle the protons into the all-important proton gradient that feeds cellular machinery. However, our particular strain, CFN42, doesn't have any capture hydrogenase. Nonetheless, since nature developed pathways to do the exact opposite of what we're doing, we were interested in finding out any toxic or deleterious effects our system might have. Therefore, out goals can be stated as being:
- Under symbiont form, our transgenic chassis is capable of Hydrogen production?
- To what extent does the Hydrogen production affect core metabolism, and Nitrogen fixation?
- Is there a toxic effect derived from Hydrogen production?
Theoretical Background
FBA is part of a larger set of models known as the Constraint Based Analysis (CBA). Since cells may adopt an astronomical level of possible solutions for a given set of conditions, CBM models create constrains on the solution space to limit the complexity of the solution and render it easier to compute, interpret, and/or understand.
FBA starts at the assumption that pathways strive towards homeostasis. Thus, it assumes the cellular chemoton will adjust whatever it has to do in order to maintain chemical stability. In other words, this model does not require pesky kinetic constants, something that made our lives easier. For some light reading on this, you can consult "Systems Biology: Properties of Reconstructed Networks", by Bernhard Palsson. Or you can always go ask the All-Knowing-Oracle here.
The Simulation
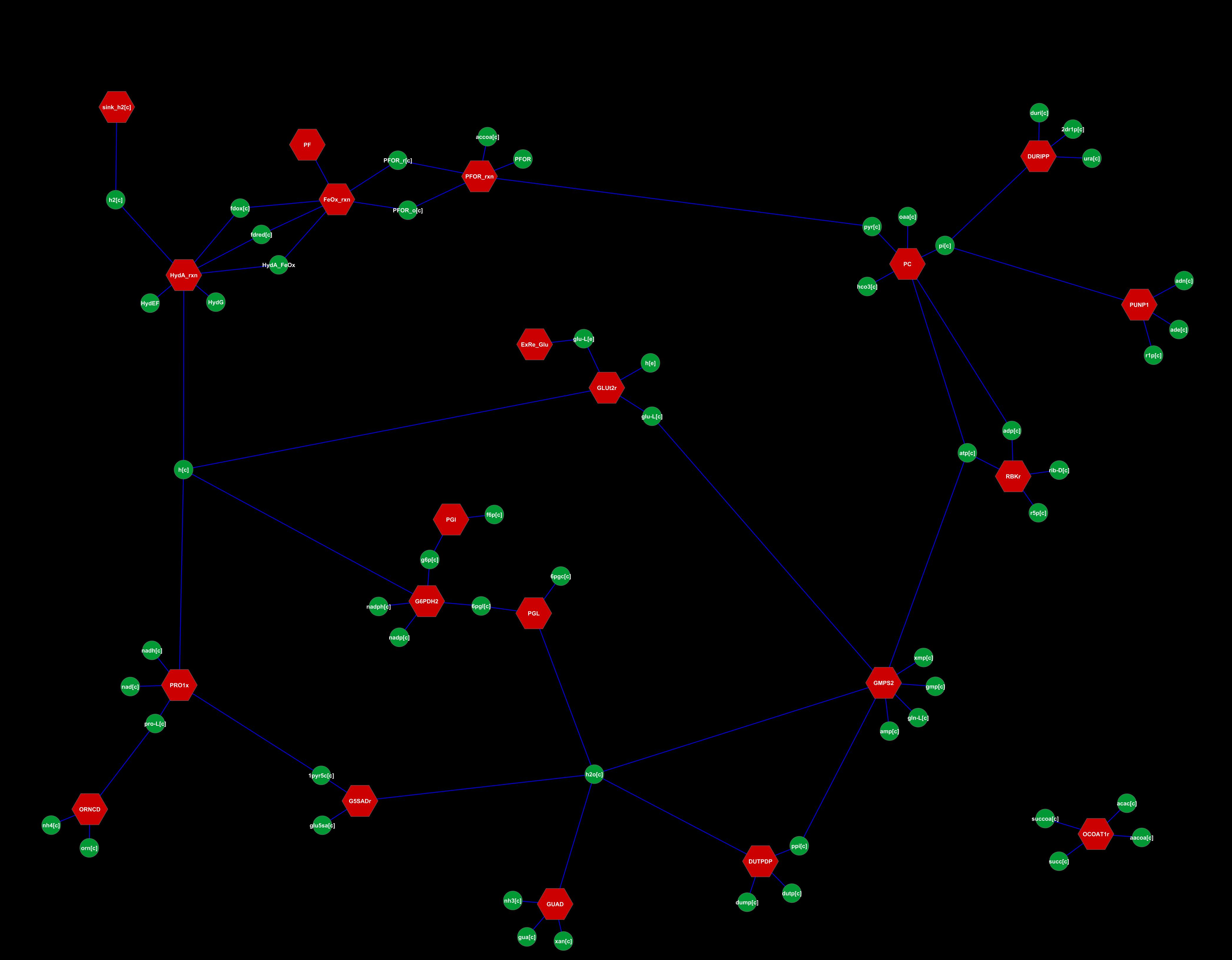
A researcher at our institute had already done a metabolic reconstruction for the core metabolism of our chassis under symbiont form. The proteins in this reconstruction were chosen based on microArray data, and the pathways were reconstructed using the then-available literature. The data looks like:
The Toolbox
We performed the FBA calculations using a particular toolbox in iGEM's favorite program: MATLAB. In particular, we used the COnstrained Based Reconstruction & Analysis toolbox (COBRA), originally by the Palsson Lab. I have to confess, the poor documentation (version 3.1.2) made our life difficult. It appears the toolbox has a lot of functions for various CBA, though a simple tutorial would have been quite welcome...
Parsing v1
At first we decided to parse the data we had into The Stochiometric Matrix, and we did it, with a combination of PERL, R, & MATLAB scripts. We had a lot of trouble with that parser particularly because we didn't know to what structure it had to conform (i.e. what are the fields of "rev", or "c", or "b"...). After finding this sample file to use and the Nature Method Tutorial, we eventually got it working. Once the thing was partially functional, we got to play around with COBRA's functions, and we found the elusive createModel function.
Parsing v2
With the createModel function, we re-made the COBRA structure and got it fully functional with this. It was then we discovered the data we had been working with was painfully out-of-date. Searching in vain for a magical function that would consult databases and update reactions, we realized manual curation was necessary. For every one of the 385 reactions we had.
Parsing v3
We used the E-C Numbers classification to check the known reactions, we connected isolated dots with the Kyoto Encyclopedia of Genes and Genomes, and we finished the holes with other searches... Once the data felt up-to-date, we set to re-analyze and tweak the metabolic reconstruction to our heart's content with this. We made several models to explore how the system reacted:
| Model | Core Metabolism | H2 Sink | H2 Production | PFOR activity | FeOx activity | PHB | File |
|---|---|---|---|---|---|---|---|
| WT Model | Yes | No | No | No | No | No | WT Model |
| TG Model: Main | Yes | Yes | Yes | Yes | Yes | Yes | TG Model Main |
| TG Model: sub 1 | Yes | No | Yes | Yes | Yes | Yes | TG Model 1 |
| TG Model: sub 2 | Yes | No | No | Yes | Yes | Yes | TG Model 2 |
| TG Model: sub 3 | Yes | No | No | No | Yes | Yes | TG Model 3 |
| TG Model: sub 4 | Yes | No | No | No | No | Yes | TG Model 4 |
| TG Model: sub 5 | Yes | Yes | Yes | Yes | Yes | No | TG Model 5 |
We do realize there's an option to knock-out a gene, and then perform the analysis, but this approached yielded easier-to-interpret results (in our humble opinion). We kept the Objective Function (OF) of the old data since it made sense. After running all the models, we realized that our OF was maximized for the Main Transgenic Model.
Parsing v4
Since our pathway was adding a novel metabolite, we hypothesized the transgenic pathway might represent an Extreme Pathway (you know, the regular hardcore, skydiver, shark-wrestler, wompa-tamer, lightsaber-juggler kinda of pathway...). For all the nasty details, go here. Anyhow, getting the data into the format acceptable by ExPa required another round of parsing, and another script.
Visualizing
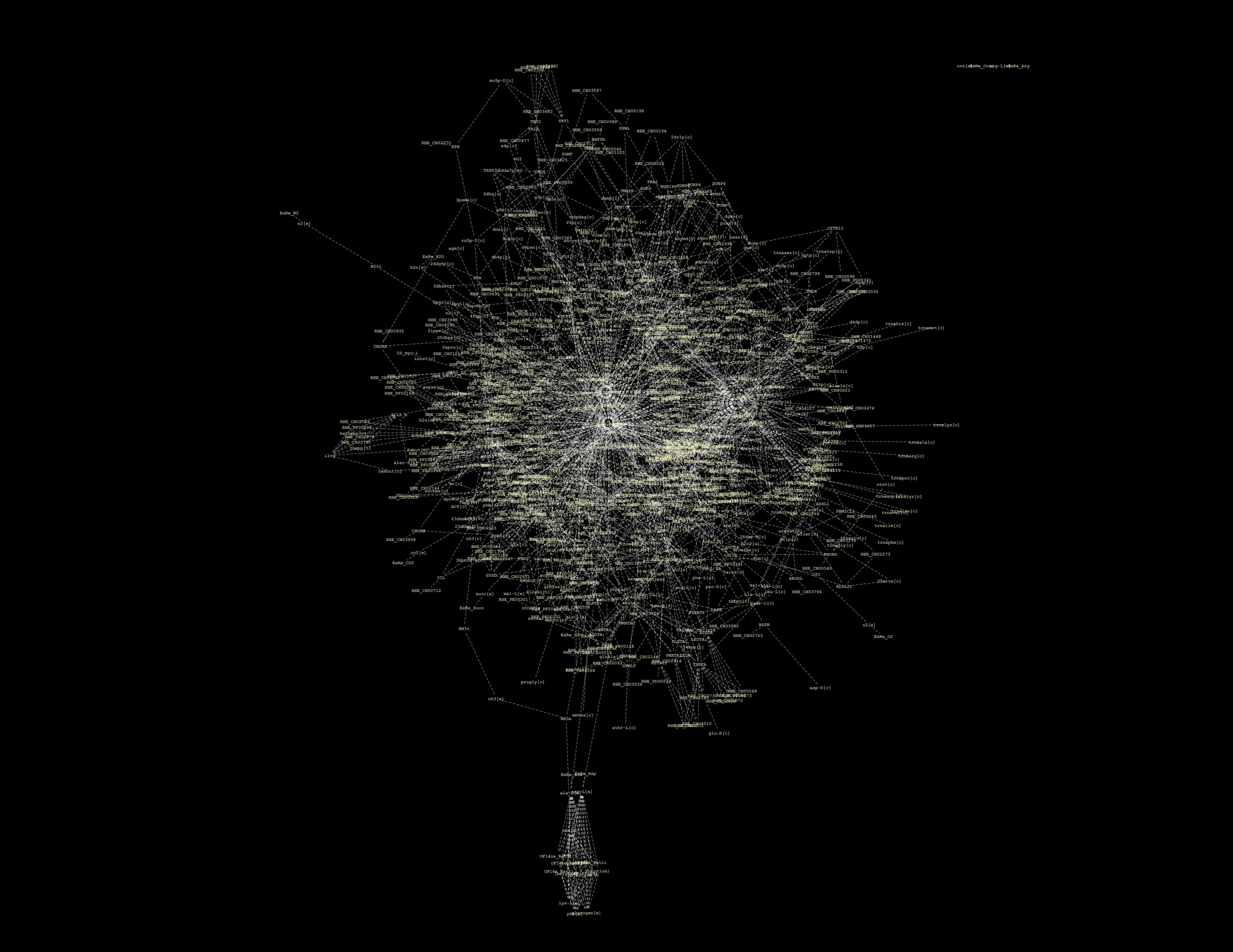
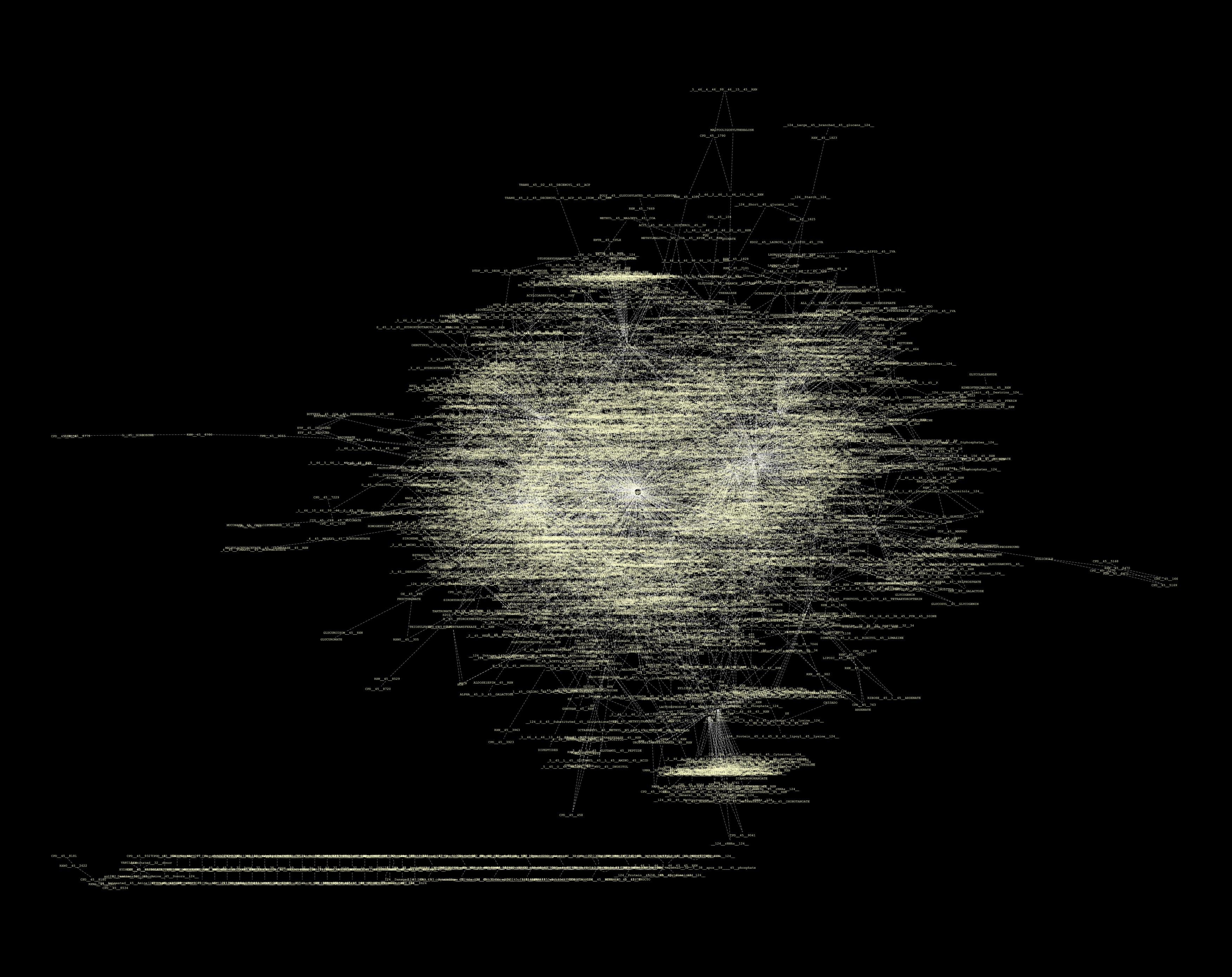
In all this, we have been looking for a decent way to present our findings. However, the full network for the whole metabolome is just to massive to represent in a visually meaningful way:
We've tried Arcadia, but we get strange errors (apparently, libSBML errors, again). We've tried Cytoscape, but again the network is just so massive it is impossible to work with (in our humble personal computers that is). We tried the Edinburgh Pathway Editor, but it was not that functional to our problem as the Import Options were non-existant. We also tried SBMM but got the weird "HTTP Status 500" error. Therefore, we opted for two approaches : fraction our data into modules, sorted by "sub-compartment" (i.e. metabolic classification); and work with only the reactions changing between WildType and TransGenic strains.
A note on the Edinburgh Pathway Editor, while it is extremely unstable and crashes more often than BethSoft's Third Scroll (beware and abuse the Save option), it is functional enough to create pathways and illustrations. The main image at the Introduction Section was created using said Editor.
The Results
So, what have we learnt form all this parsing around? Well:
- Under symbiont form, our transgenic chassis is capable of Hydrogen production?
- Flow through the Hydrogen output reaction is maximal for our TG model, therefore it does produce Hydrogen.
- To what extent does the Hydrogen production affect core metabolism, and Nitrogen fixation?
- Nitrogen fixation is increased to 150%, we believe we are detoxifing the chassis from redox potentials (see note below).
- Carbon intake is increased to 130%, we believe our system require more energy to compensate redox export (see note below).
- Is there a toxic effect derived from Hydrogen production?
- The Objective Function remained virtually un-altered, therefore the chassis' fitness is virtually unaffected. While it has a less efficient carbon metabolism, it is a more effective nitrogen fixator.
In fact, our chassis remains quite similar to the wild type chassis (at least metabolically speaking...), to the point where there are only around 20 different reactions between the two (16 without the 4 reactions that make up the H2 pathway). These 16 reactions for the TransGenic are beyond one order of magnitude different from their WildType counterparts. As a side note, when we used a >1% difference decision threshold, we found over 105 reactions. However, we believe that the order of magnitude logic is more relevant to a biological context of metabolic reactions. Therefore, our final networks looks like this:
Our synthetic pathway appears in the top left section. The center holds carbon metabolism, and the periphery has some miscellaneous reactions...
Note: We believe thus due to the results of our different models. While the Main model produced maximal hydrogen and maximal OF, the rest of the submodels produced markedly less hydrogen and OF. The only way to restore maximal efficiency was to re-introduce the H2 sink, that is to remove H2 from the system. The key point in this is the removal of something from the chassis. Since H2 is made up of 2 protons and 2 electrons (or reduced H+) that species had to be either protons or electrons. Since Nitrogen fixation remained very high, and said reaction consumes protons, the mystery species had to be electrons. In other words, redox potentials.
References
Someday, after someone installs the Cite/Cite.php extension on this server, the <references /> tag will work. Until then, references appear inline...
 "
"