Team:UEA-JIC Norwich/Nittygritty-algae
From 2011.igem.org
Benjevans1 (Talk | contribs) |
Benjevans1 (Talk | contribs) |
||
| Line 121: | Line 121: | ||
<p>We are planning to transform the algal species <i>Chlamydomonas reinhardtii</i>. This is a single celled, photosynthetic eukaryote. It is easily transformable, either by: electroporation; the bacterium Agrobacterium tumorfaciens; glass beads; or by the use of a biolistic particle delivery system (gene gun). We will be using the specific strain CC-4350 cw15-302 mt+. This strain is biflagellate with a high transformation frequency, due in part to its lack of a cell wall. Its genome has been sequenced, allowing us to research its codon bias.</p> | <p>We are planning to transform the algal species <i>Chlamydomonas reinhardtii</i>. This is a single celled, photosynthetic eukaryote. It is easily transformable, either by: electroporation; the bacterium Agrobacterium tumorfaciens; glass beads; or by the use of a biolistic particle delivery system (gene gun). We will be using the specific strain CC-4350 cw15-302 mt+. This strain is biflagellate with a high transformation frequency, due in part to its lack of a cell wall. Its genome has been sequenced, allowing us to research its codon bias.</p> | ||
<p>There are twenty common amino acids, but over sixty codon configurations. So, each amino acid can be coded for by multiple codons. The code is thus said to be degenerate. However, most organisms display a preference for one codon or another, and so prefer to express a given amino acid by a certain codon. This is known as the codon bias. We researched the codon bias for <i>Chlamydomonas reinhardtii</i>, and plan to use this information to optimise the Biobricks we wish to use for expression in <i>C. reinhardtii</i>. | <p>There are twenty common amino acids, but over sixty codon configurations. So, each amino acid can be coded for by multiple codons. The code is thus said to be degenerate. However, most organisms display a preference for one codon or another, and so prefer to express a given amino acid by a certain codon. This is known as the codon bias. We researched the codon bias for <i>Chlamydomonas reinhardtii</i>, and plan to use this information to optimise the Biobricks we wish to use for expression in <i>C. reinhardtii</i>. | ||
| + | </html> | ||
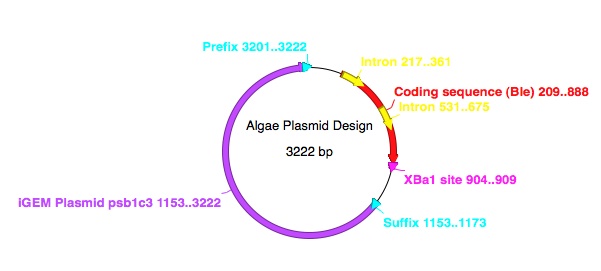
| + | [[File:Circular_plasmid_algae.jpg]] | ||
Revision as of 15:47, 18 July 2011
Algae
We are planning to transform the algal species Chlamydomonas reinhardtii. This is a single celled, photosynthetic eukaryote. It is easily transformable, either by: electroporation; the bacterium Agrobacterium tumorfaciens; glass beads; or by the use of a biolistic particle delivery system (gene gun). We will be using the specific strain CC-4350 cw15-302 mt+. This strain is biflagellate with a high transformation frequency, due in part to its lack of a cell wall. Its genome has been sequenced, allowing us to research its codon bias.
There are twenty common amino acids, but over sixty codon configurations. So, each amino acid can be coded for by multiple codons. The code is thus said to be degenerate. However, most organisms display a preference for one codon or another, and so prefer to express a given amino acid by a certain codon. This is known as the codon bias. We researched the codon bias for Chlamydomonas reinhardtii, and plan to use this information to optimise the Biobricks we wish to use for expression in C. reinhardtii.
 "
"