Team:UC Davis/Notebook wip
From 2011.igem.org
Start a Family
Got a favorite BioBrick? Check our our process for expanding basic parts into part families.Criteria
View our judging criteria for iGEM 2011 here.
Contents |
Week 1
--Monday 6/13/11--
Today we had our first real day of lab work. We settled ourselves into the lab bench that we will work at for the summer and set up our area. Along with familiarizing the new members with lab protocols, we had a productive day of hydrating and transforming. We really liked the linearized vectors that the Registry sent along with the plates, very nice addition. Today we designed what our wiki will look like and talked about some finer of the details of our project such as the exact construction method and a work plan. We are considering using Gibson assembly for all of our construction although in our lab it is still being debugged in a different side project. Results are promising though as biobricking has its drawbacks. Among other things, planning of an all-you-can eat sushi lunch is in the works. We will plan our work schedule around that.
-Rehydrated parts from 2011 Registry Distribution:
- -R0040=Tet promoter in psb1a2
- -C0040=Tet repressor in psb1a2
- -C0012=LacI repressor in psb1a2
- -C0051=Lambda cI repressor in psb1a2
- -R0051=cI-regulated promoter in psb1a2
- -I732006=LacZ-alpha in psb1ak3
- -R0010=LacI regulated promoter in psb1a2
- -E0040=GFP coding region in psb1a2
- -J23101=Constitutive promoter in j61002
- -C0080=AraC repressor/activator in psb2k3
- -I13458=pC+AraC in psb1a3
- -I13453=pBAD promoter in psb1a3
- -B0015=Double terminator in psb1ak3
-Transformed all of these hydrated parts
--Tuesday 6/14/11--
Today we tested out error-prone PCR on the LacI promoter. We are adding MnCl2 to a few of the tubes and changing the nucleotide concentrations in other tubes. We want to test out some different screening techniques for a rapid hunt. Characterizing thousands, possibly tens of thousands of promoter mutants is quite a daunting task and we want to make it as easy as possible. We are looking to using a fluorescent cell sorter on campus which could narrow down a few good candidates for more rigorous testing.
We began work on the actual wiki with some code that I (Tim) wrote over the weekend. It works perfectly in an unrestricted webpage but once I put it in iGEM's wiki, it doesn't work. We'll have to figure this out. We cultured all parts and digested to check on a gel. All parts checked out. Sent out all hydrated parts to get sequenced.
We ligated mutants into screening plasmid to see if our error-prone PCRs had visible mutants.
Additionally, we rehydrated and transformed part C0080 (araC regulatory protein) as we had forgotten to rehydrate it on Monday.
--Wednesday 6/15/11--
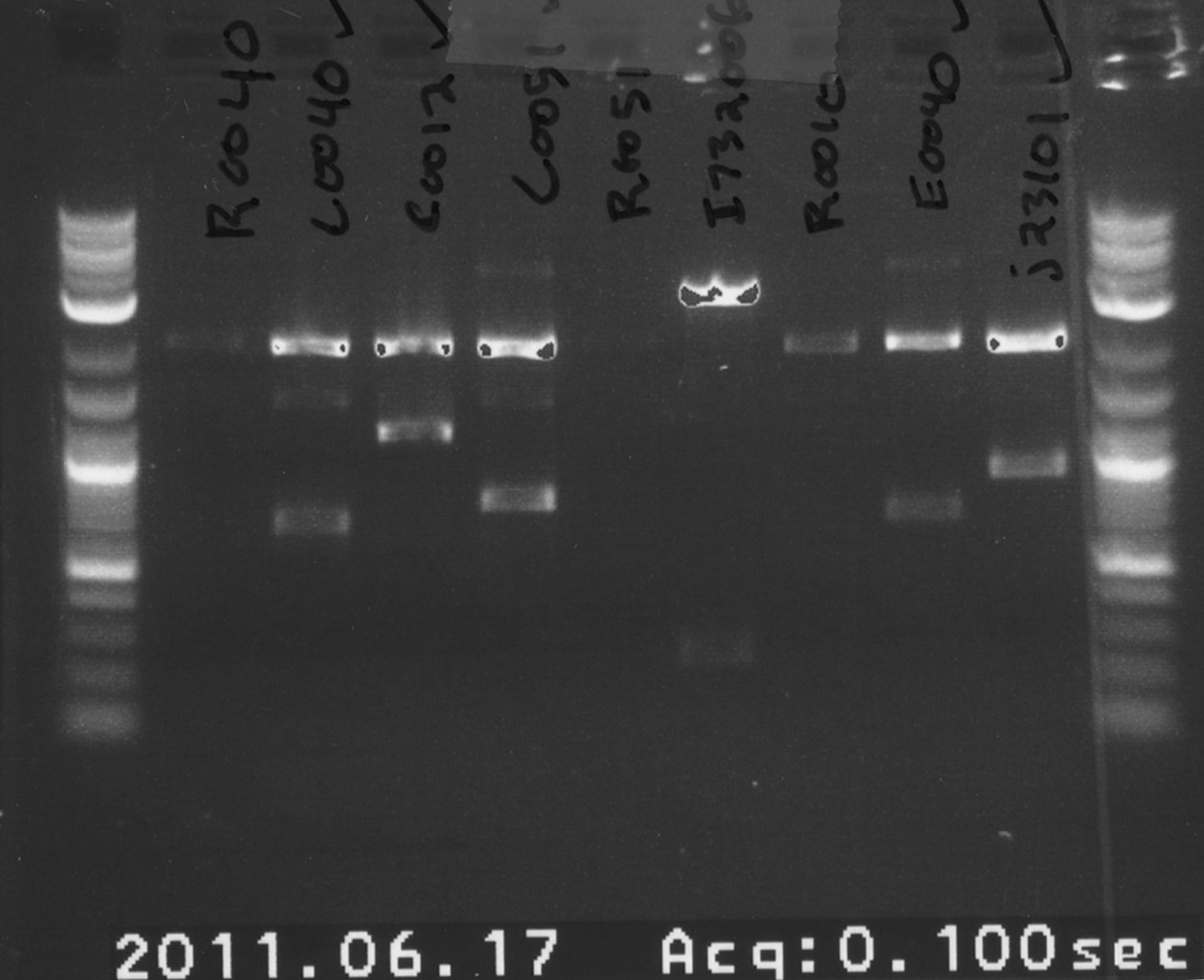
Today we ordered primers for mutagenesis of LacI promoter, cI promoter and tet promoter. They didn't have adequate overhangs on the forward primer which could make it difficult or possibly impossible to digest in order to put in a screening plasmid. We also ordered primers to replace RBS (Bba_B0032) in Bba_E0240 to Bba_B0034. Glycerol stocks were made of our hydrated parts. We sent all of our parts in to get sequenced to avoid problems we've had in the past with incorrect things from the Registry. We miniprepped and digested the parts that were cultured in order to do a rough check on a gel while we're waiting for our sequencing data to come back.
We miniprepped and digested the following parts with XbaI and PstI:
- -R0040
- -C0040
- -C0012
- -C0051
- -R0051
- -I732006
- -R0010
- -E0040
- -J23101
Additionally, GFP (E0040) was digested with EcoRI and SpeI.
--Thursday 6/16/11--
Today was an exciting day! We went down to Stanford for a meetup with iGEM teams from UCSF, UC Berkeley, and Standford/Brown. With the little time we had before our road trip, we were able to miniprep C0080 and digest the mutant LacI promoter. Once at Stanford, we discussed our projects and our strategies for approaching them and talked with each other to share information that could be valuable to each other's projects. The UCSF team is very impressive being mostly comprised of high school students who seem very sharp. After a bit of mingling, we all headed to a different part of campus where SB5.0 was happening. We were lucky enough to be able to attend the poster session which was great! There were so many posters which all presented interesting things that were similar to things we have worked on or were interested in working on. A few notable posters showed mutating one domain in an efflux pump that were aimed at pumping out biofuel. One poster which we were interested in was one that raised the idea that DNA could be made with a different backbone which could possibly introduce another mode of isolating synthetic systems in a complex cell. No doubt there would be a boatload of work involved with building artificial machinery to do the basic functions which have evolved for millions of years in nature. Perusing the posters gave us some ideas and really motivated us to do work on our project.
--Friday 6/17/11--
Today we did some transformations of Bba_E0240 ligated to our magic screening plasmid. Those plates will be taken out on Saturday morning and cultured Sunday evening. We also ran our digestions on a gel to check the lengths. The mostly came out well. Some of the promoters were very faint and a bit inconclusive but we'll rerun those later.
Sushi buffet today. Fullness has taken on a new meaning.
--Saturday--
Today Tim took out the transformation plates and put them in the 4 degree room for storage until tomorrow.
--Sunday--
Today Keegan cultured all the transformants in order to miniprep them on Monday.
Week 2
--Monday 6/20/11--
Today we nanodropped the 12 digested mutated LacI promoters and the J61002 promoter screening plasmid such that we could ligate them:
| Sample ID | ng/uL | 260/280 |
| LacI 1 | 188.93 | 1.19 |
| LacI 2 | 113.96 | 1.26 |
| LacI 3 | 87.29 | 1.19 |
| LacI 4 | 88.23 | 1.20 |
| LacI 5 | 109.35 | 1.22 |
| LacI 6 | 90.64 | 1.18 |
| LacI 7 | 536.16 | 1.33 |
| LacI 8 | 415.75 | 1.33 |
| LacI 9 | 462.50 | 1.33 |
| LacI 10 | 544.12 | 1.36 |
| LacI 11 | 461.32 | 1.33 |
| LacI 12 | 498.83 | 1.35 |
| J61002 | 11.92 | 1.95 |
We then ligated the 12 mutated LacI promoters into the J61002 promoter screening plasmid. We had planned to transform all of these ligations but unfortunately ran out of LB+Carb plates. We were still able to transform about 7 of the 12 ligations. We also rehydrated and transformed the B0015 terminator.
We miniprepped and digested the following parts:
- I13458 (with EcoRI and SpeI)
- I13453 (with EcoRI and XbaI)
- R0010 (with SpeI and PstI)
- R0040 (with SpeI and PstI)
- R0051 (with SpeI and PstI)
--Tuesday 6/21/11--
After yesterday's plate shortage, we took on the new task of making new LB+Carb plates.
We also nanodropped many of the minipreps (MP) and gel purifications (GP) from earlier:
| Sample ID | ng/uL | 260/280 |
| J23101 MP | 338.75 | 1.87 |
| I13456 MP | 245.92 | 1.86 |
| I13453 MP | 181.48 | 1.85 |
| R0010 MP | 115.98 | 1.86 |
| R0051 MP | 101.21 | 1.89 |
| E0040 MP | 240.71 | 1.82 |
| R0040 MP | 80.18 | 1.92 |
| C0012 MP | 165.16 | 1.87 |
| R0010 GP | 22.42 | 1.73 |
| I13458 GP | 16.63 | 1.86 |
| R0040 GP | 20.39 | 1.62 |
| I13453 GP | 15.10 | 1.74 |
| R0051 GP | 7.27 | 2.69 |
--Wednesday 6/22/11--
Miniprepped B0015 and made glycerol stocks of I13453, I13458, and B0015. We also made some 10 mM IPTG from more concentrated stocks.
--Thursday 6/23/11--
After waiting most of the week some of our primers finally came in! We ran PCR with our B0034+E0240 forward primer and our E0240 forward primer.
--Friday 6/24/11--
PCR purified B0034+E0240 and E0240. Digested B0034+E0240 with XbaI and PstI.
Today we digested the following parts:
- B0034 (with SpeI and PstI)
- C0040 (with XbaI and PstI)
- C0051 (with XbaI and PstI)
- C0012 (with XbaI and PstI)
- J61002 (with EcoRI and PstI)
- psB1K3 (with EcoRI and PstI)
Week 3
--Monday--
Today we made Dh5a competent cells as we had run out of our stocks. Keegan made a Gibson volume calculator which will easily calculate the volumes of each part you need when you input the concentrations of those parts to make sure you have equal molar amounts. It will be handy for quick calculations of assemblies of up to 10 parts at a time.
--Tuesday--
Today we transformed the LacI promoter mutants into the screening plasmid Bba_J61002. We're hoping to see red from the RFP that is built into the screening plasmid. Ideally we will see shades of red corresponding to differing strengths of the mutant promoters. Although we're planning on using a fluorescent cell sorter, a cursory screening could be useful just to make sure our error-prone PCR was successful. We transformed into Dh5a E. coli so we had to induce our plates with IPTG since this particular strain is LacI+.
We had some very unusual weather here today. Normal Davis summers are very hot and dry and today should have been around 90-100F but it was rainy and cold! Global climate change perhaps?
--Wednesday--
We realized today that we needed to test some carb plates that we made last week. Our suspicions arose when 16 ligations that had previously been transformed successfully failed to grow on our new plates. We did a quick test by streaking a couple plates with a part in an AK backbone. We streaked the same part on both Kan and Carb plates and only the Kan plates had any growth on them. We will make some new Carb plates tomorrow and hopefully they'll work! Today we also designed the rest of our Gibson assembly primers for constructing our screening plasmid. We'll order tomorrow and in about a week be able to assemble everything fairly quickly. In the meantime we'll continue using biobrick assembly.
--Thursday-- We did PCR of E0240 with and without B0034. We wrote our formal letter to Novozymes and set up digest with E and P of E0240 modified with and without B0034. We also got a new strain of E. coli, BW22826 which is F-, Δ(araD-araB)567, Δ(codB-lacI)3, &lambda-, rpoS396(Am), rph-1. This strain will be helpful for us since it is both araBAD and LacI negative. This means we won't have to put an arabinose repressor in our plasmid which will cut down on construction time and allow for more testing time! We also needed the strain to be ∆LacI since we will be testing the Lac repressor and operator.
We also made carbenicillin plates and diagnosed the "bad" carb plates as chloramphenocol plates. Our mistake came when we used an older stock of carb which was labelled only with a 'c'. Lesson learned; don't assume anything!
--Friday--
Transformed all of our LacI mutants again along with E0240+psb1k3. All of the LacI mutants were plated on newly made Carb plates. Hopefully they have the correct antibiotic this time! We also got a new strain, JS006, which we requested from another lab. We'll make glycerol stocks and competent cells of these.
--Saturday-- Took plates out and all had growth=carb plates good! It's amazing what plating on the right antibiotic will do. All of our plates which had failed just a few days ago had colonies with a few of them being red.
Week 4
--Monday 7/04/11--
Happy 4th of July for all the Americans! Today we started making more competent cells for the new strain BW22826. Tomorrow we will complete the protocol.
--Tuesday 7/05/11--
BW22826 Cells had very fast growth overnight, and were too overgrown to use for competent cells in the morning. The procedure was restarted late at night (around 9:30) and lesser amounts of starter culture were used (1, 2, and 2 mL) to ensure the cells don't overgrow. One culture was placed on a shaker at room temperature, the other two were placed on a bench at room temperature.
We inspected our LacI mutants. Most of the plates were covered in white colonies, though a few had some slightly pink colonies, indicating mutations were successful. The cells were plated on IPTG+ plates, so we expect induction; it is possible that the mutation rate was high and the promoter function was destroyed in a majority of colonies. Ten pink and ten white colonies were replated on IPTG+ plates, to rule out poor induction on our initial plates.
E0240 variants with B0034 and with no RBS in pSB1K3 were cultured.
--Wednesday 7/06/11--
The making of our competent cells was successful! We were able to produce two full boxes today, with good amounts of cells in each of the tubes. Hopefully we won't have to make more for a while.
We also miniprepped the E0240 variants from yesterday so that we could proceed with construction. We digested these minipreps along with our three main promoters (R0040, R0051, R0010) so that we could ligate them together.
--Thursday 7/07/11--
Today started slow but ended in a flurry! We ligated three promoters (R0010, R0040, R0051) to E0240 (GFP) and transformed them. We also retransformed our mutant LacI ligations to try and get even more. The previous batch of potential mutants which were different shades of red on a replica plate got replated to see if we'll be able to see a clearer difference between shades of red on streaked plates. So far there appears to be a good range of redness which is a positive sign that our mutagenic pcr worked somewhat well. Out of 12 plates, we got 9 possible mutants. We're planning on taking a closer look with our Tecan plate reader early next week. This will give some more definitive proof of good mutants or not. These mutants also got cultured so we can prepare them for sequencing soon.
In the Dh5α strain, LacI repressor is always present so, upon induction we expected to see red. It seems from our mutation protocol that we mutated them too heavily since most colonies were white even with IPTG present.
--Friday 7/08/11--
Mutated C0012, C0051, C0040. Ligations of promoters to GFP looked green=good! Made control constructs for R0010 Ligated R0040 and R0051 mutants into promoter screening plasmid for screening!
Week 5
--Friday 7/15/11-- Today was a really fun day! We did some minipreps in the morning but then drove down to UC Berkeley for a NorCal iGEM picnic. We got to talk with our fellow iGEMers from UCSF and Berkeley and got updates on how their projects were going. It was nice to see that there is a community of synthetic biologists in Northern California that we can share ideas and materials with. After a relaxed lunch, we had a small meeting to discuss how to achieve the medal requirements. We discussed helping each other to fulfill one of the gold medal requirements with the Berkeley team since some of our software tools might be useful for their project this year. We then got to see their lab and received some pBAD+AraC since those parts from the registry seem to be faulty. Overall a really good day!
Week 6
Week 7
Week 8
Week 9
Week 10
</html>
 "
"