Team:TzuChiU Formosa/Notebook
From 2011.igem.org
(→2011.09.10) |
(→Photopaper) |
||
| Line 101: | Line 101: | ||
==='''2011.09.07'''=== | ==='''2011.09.07'''=== | ||
<pre> | <pre> | ||
| - | ''' | + | '''Rhodobacter rudrum medium''' |
K2HPO4 1g | K2HPO4 1g | ||
NaCl 0.5g | NaCl 0.5g | ||
| Line 131: | Line 131: | ||
File:01.jpg | File:01.jpg | ||
</gallery> | </gallery> | ||
| + | |||
==='''2011.09.08'''=== | ==='''2011.09.08'''=== | ||
| Line 186: | Line 187: | ||
→37℃ for 30 mins | →37℃ for 30 mins | ||
</pre> | </pre> | ||
| + | |||
==='''2011.09.11'''=== | ==='''2011.09.11'''=== | ||
| Line 205: | Line 207: | ||
PSB1C3 backbone | PSB1C3 backbone | ||
</pre> | </pre> | ||
| + | <gallery> | ||
| + | File:02.jpg | ||
| + | File:03.jpg | ||
| + | </gallery> | ||
Revision as of 08:55, 18 September 2011


Photopaper
2011.02.24
Discussion:
- Team organization
- Brain storming
- paper made by bacteria with add-ons such as colors, fragrance, etc.
- "light up" the plants for replacing lamp posts.
2011.03.04
Discussion:
- Team advisory
- Brain storming
- Add-ons: colorful cellulose which produced by bacteria such as beta-carotene
- information exchange with iGEM 2009 Cambridge team
2011.03.14
Discussion:
- Task Allocation
- Brain storming
- Avatar - "light up" the plants by transfecting symbiotic bacteria which cloned into fluorescence gene
- Eco-friendly warmer - biotic thermal pad
2011.03.23
Discussion:
- Project : paperia
- Option 1 : Culture bacteria which has pigment gene
- Option 2 : Cellulose-producing bacteria secrete pigment into the medium
2011.03.24
Discussion:
- Exp. procedure:
- cloning of cellulose gene’s CDS
- the product should operate within E. coli.
2011.06.22
Discussion:
- Due to some unforseen reason, the team decided to change their project.
- New project: Biojenny
- -economical and humane way to produce paper in large quantities.
- -yeast to be our host
2011.07.01
Discussion:
- Freeze > grin > genome DNA isolation > Cloning = silk protein gene
2011.07.09
Discussion:
- the connections between 3 silk proteins : Fibl Fibh P25
- major proteins : H-chain, L-chain, P25
2011.07.15
Discussion:
- Due to limited resources and techniques required, the team decided to switch back to the previous project. Paper making !
- However it would be modified to be more innovative and creative.
2011.07.18
Discussion:
- Latest project : Photo paper
- cyanobacteria is designed as the host, cellulose made up of the glucose produced by cyanobacteria could be one of the main attraction of the project.
2011.07.23
Discussion:
- system modification to overcome the problems arises during preliminary round
- Biobricks from Tokyo 2010 team will be utilized
- regulator promoter in order to regulate the secretion of cellulose to solve the aggregation of the bacteria
2011.09.07
'''Rhodobacter rudrum medium'''
K2HPO4 1g
NaCl 0.5g
FeSO4.7H2O 0.01g
CaCl2 0.02g
MnCl2.4H2O 0.002g
MgSO4.7H2O 0.2g
NaMO2O4.2H2O 0.01g
ddH2O 998.258 ml
(+
______________________________________________________
1L→take100ml
+
Yeast Extrat 0.5g
Sodium malate
(Sodium succinate dibasic hexohydrate) 5g
NH4Cl 1g
ddH2O 893.5ml
(+
______________________________________________________
1L
'''Raise E. coli(PSB1C3)'''
1.50ml LB+500μl CHLORAMPHENICOL
2. 37℃, overnight (14-16hrs)
2011.09.08
'''Plasmid miniprep kit''' PSB1C3 plasmid '''revive Rhodobacter rubrum'''
2011.09.09
'''Raise Gluconacetobacter hansenii''' 1.50ml LB+500μl CHLORAMPHENICOL 2. 37℃, overnight (14-16hrs)
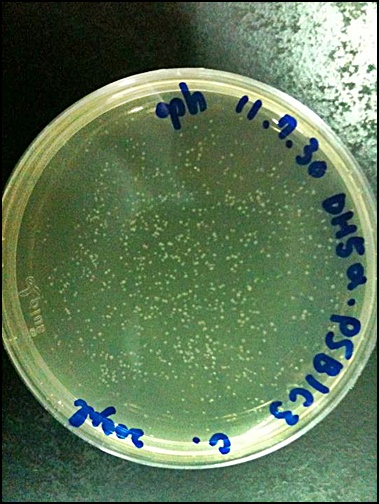
2011.09.10
'''Digestion check of DNA'''
PSB1C3 DNA (control) 3μl
PSB1C3/ EcoR
DNA 10μl
10×buffer 5μl
BSA 5μl
EcoRⅠ 1μl
ddH2O 29μl
(+
______________________________________________________
50μl
PSB1C3/ pstⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
pstⅠ 1μl
ddH2O 29μl
(+
______________________________________________________
50μl
PSB1C3/ EcoRⅠ+pstⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
EcoRⅠ 1μl
pstⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 30 mins
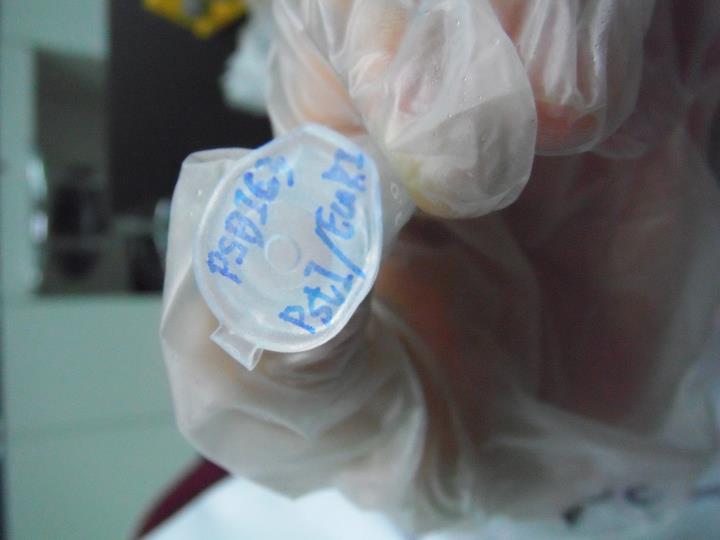
2011.09.11
'''Digestion of DNA'''
PSB1C3/ EcoRⅠ+pstⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
EcoRⅠ 1μl
pstⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 2 hrs
'''electroelution Purification'''
PSB1C3 backbone
2011.09.13
'''Raise Rhodobacter rubrum''' 1.50ml LB+500μl CHLORAMPHENICOL 2.37℃, overnight (14-16hrs)
2011.09.14-17
'''PCR'''
template (Gluconacetobacter hansenii) 180μl
5×Buffer 144μl
2.5μM dNTP 14.4μl
10μM F 36μl
10μM R 36μl
phusion DNA (polymerase) 7.2μl
ddH2O 302.4μl
(+
______________________________________________________
720μl
2011.09.15
'''Genome miniprep''' Gluconacetobacter hansenii
2011.09.16-17
Gluconacetobacter hansenii
File:.jpg </pre>
CO killer
2011.07.04
Today we had our meeting in the afternoon. The weather is so hot. It was like barbeque ourselves under the sun so we decided not to have meeting in the afternoon anymore. The topic today is the oxidation of CO to CO2 – we found out that Rhodospirillum rubrum can undergo Calvin Cycle to obtain their energy from sunlight and use CO2 as their carbon source, however cyanobacteria or other bacteria is needed for CO2 fixation and production of biofuels. But it would be much more sophiscated if we add on the cyanobacteria into the system. Hmmm … looks like we have to improve more on our system design !
2011.07.05
We talked to the microbiologist Prof. Chen CY yesterday evening and stunned us with 3 questions. The time factor, the mutation factor and the gene expression rate. The experiment of CO2 fixation requires at least half a year to complete so we probably couldn’t just simply add in our project. Besides that, we also have to think of the mutation of the gene. If the gene mutated then the product would be useless. And the most important is the rate of the protein to turn fluorescent. Lastly, if the gene size is getting bigger it would be more difficult to insert into the plasmid. Consequently, we would need more time in cloning and that would be time-costing. Uhh … we really need to find the resolution soon ! Any way, Prof. Ingrid is really very busy. We have not see her for the past 2 weeks. Gotta check out with her lab assistant.
2011.07.06
Today we had meeting in the afternoon again. Thanks god the weather is windy. We downloaded all the sequences and CDS of the gene we need. On the other hand we also search the sources of R. Rubrum which we planned to use in our project. There is a few proffesor in taiwan doing the research about R.Rubrum. So Joanne will help the letter to them to request a sample of it. And the worst case we can buy it from biotech corporation. Steven need somebody to be the in-charge of wikipedia. Leo volunteered. Found out that 2 enzymes are available : CODH–Nickel, OM5 –Molybdenum; CODH is has a higher rate of binding with CO, but OM5 can bind with CO in enormous amount at one time. Say no to procrastination ! Next meeting on Friday. A day off tomorrow =D
2011.07.09
Discussion regarding the presentation slide during the preliminary round. Task allocated to all the members. While Joanne and Steven had to write the project proposal ASAP. Not much were done today. But TGIF !! Had some photo session after the meeting. Joanne go crazy with the effect thingy on the MacBook. Stressed up after one week of intensive meeting.
2011.09.08
'''Raise E. coli(K325903&K325229) from CAMBRIDGE''' 1.50ml LB+500μl CHLORAMPHENICOL 2.37℃, overnight (14-16hrs) 3.L-arabinose test Neither of the two E. coli is feasible.
Helmersi Codon
2011.02.13
Let the participants to propose some ideas:
- To make the waste become ink after bacterial reaction.
- Make the bacteria to produce combustible materials.
- Make the bacteria to emit different light.
- Edible bacteria.
- To control the shape of bacterial.
- To make the bacteria a fat eater.
- Glowing mushrooms.
- Make the bacteria to stick with dust.
Poll result:
Top three projects:
- Edible bacteria.
- Glowing mushrooms.
- Combustible bacteria.
Conclusion:
Let all participants to choose their project and make a group then check the extent of implementation.
2011.03.12
Discuss the problem between the team members:
- The rule of conference:
- We can choose a chairman.
- We can take turns by seat number then everyone has a chance to practice how to speak stable on the stage.
Group report & Vote:
Edible bacteria had become our determined project.
Conclusion:
To make three groups and search the information of soy protein, milk protein and yeast.
2011.03.21
Group report:
- Soy protein: There have people inject a piece of soy protein, but can’t find out the sequence.
- The relation between milk protein and probiotics: There is both good and bad relation between milk protein and probiotics.
- Yeast:
Pai: Maybe we can search other organisms which have essential amino acids. Then we can complement the lack of essential amino acids in the yeast.
The application on the internet of iGEM:
Huang and Celia will teach other team members how to apply it.
The schedule of iGEM:
2011.05.23:Preliminary
The registration of iGEM:
- Registration fee: Give it to Celia
- Deadline: 2011.03.23 12:10-13:30
2011.04.12
Celiahas explain the competition system to other team members:
- Everybody needs to create a new idea and search its extent of implementation. Then design the experiment.
- On May 23, everyone has to introduce and explain the idea through the power point.
Discussion:
- How to handle with our original project?
- We can process this project and our own project at the same time.
- We can give the project to one of the team member.
Conclusion:
- We decided to give the project to Jin, but we still can process two projects at the same time if he needs our help.
- In order to help each other and solve the problems with our ideas, we still have discussion every week.
2011.05.06
Share the ideas:
- Sea water purification
- Purification of factory water use
- Bacteria for wound
- Glowing mushroom
Suggestion:
- Celia: Maybe you use plant can be easier to get the same purpose.
- Huang: Maybe you can use symbiotic bacteria of plant and inject the glowing gene into it.
2011.05.10-2011.05.12
Progress report:
- Huanghas changed her idea and is still searching the information.
- Tsai has an idea is about bio-battery. To create the ion concentration difference by bacteria and produce the electricity.
- Questions:
- Celia: If you can produce this project successfully, how do you convince the public to use your product?
- Huang: Do you have any certification to proof that you can make this project by yeast or bacteria?
- Questions:
- Zou has a new idea: To clean up our teeth by bacteria. She had find out the gene of decomposition of tartar and has the conception of teeth whitening.
- Ting find the element that both petroleum and oil has contained.
- Paiput out a new idea is odorless sweat. He chooses Helicobacter pylori and Proteus mirabilis as host.
- Jinfind the information of how Staphylococcus aureus maintain its electricity.
- Dorrie is still searching the experiment and being revised the idea.
- Yu has a new idea: Rust proof bacteria. She wanted to injected the gene into aerobic bacteria.
2011.05.25
Progress report:
- Jin: Because we have figured out the gene then we have to ask Pr. Woon to contact with the author of that paper and get the gene sequence.
Homework:
- To find the…
- The performance of E.coli and Lactobacillus.
- Protein purification.
- The plasmid of Lactobacillus.
- TA-cloning
- Huang: Send the paper to team members and ask Pr. Woon help us to get the sequence.
- Celia: Translate the conference record and store them.
2011.06.26
Progress report:
Experiment design:Jinwill discuss with Pr. Woon and set for the primer.
Homework:
- To find the farming methods of lactobacillus.
- To find the plasmid is used by Cambridge in 2010.
- To understand the design methods of primer.
 "
"