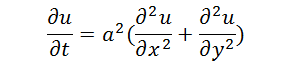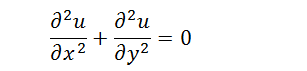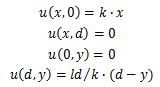Team:Tsinghua/modeling
From 2011.igem.org

Modeling
In order to depict the whole transportation process, models on formation of concentration gradient, E. Coli movement, and transcriptional regulation are essential.
Formation of concentration gradient
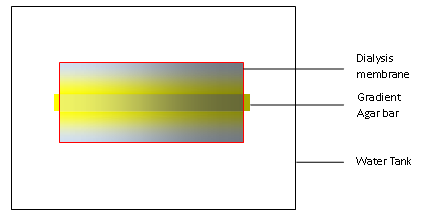
In our experiments, we first build a concentration gradient inside an agar bar as in any other traditional practice. When put in the water, the gradient agar bar will dictate the formation of a concentration gradient in water as solutes inside the agar can diffuse out. We use a dialysis membrane to wrap the bar and some water up, and then stir the water outside the membrane so that excess of solutes can diffuse outside the membrane and maintain the gradient inside the dialysis membrane.
In light of this model, it’s advisable to construct the model for gradient based on diffusion across two surfaces.
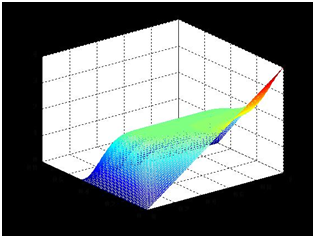
Supposing the concentration inside the bar is linear, the length of the agar bar l, and the distance from the dialysis membrane to the bar d, we might set up the coordinate with x-axis along the bar and y-axis perpendicular to the bar. The points close to the bar are on the line y=0, while points close to the dialysis membrane are on the line y=d.
When the concentration is almost steady, we shall assume that the concentration close to the bar is equal to the bar and the concentration close to the dialysis membrane is zero. Hence, concentration u
The diffusion abides by
When the system is steady, differential of u by t is zero. The equation is as follows,
The marginal conditions can be deduced as follows
Concentration plot is solved in Matlab2011a with the help of Tsinghua-A Team.
The concentration close to the bar is nearly linear, which serves as the basis for our later experiments and modeling.
E. Coli movement
It is known that movement of E. Coli is dependent on phorphorylation/dephorphorylation cycle of CheW protein and consequent shift between swimming forward and tumbling. Based on this mechanism, movement of E. Coli is largely a random process, the probability of which is modulated by certain chemicals.
Assuming that movement of E. Coli is independent of each other, we propose that the probability for E. Coli to change its direction is directly proportional to the concentration of the chemoattractant around while the choice of direction is totally random.
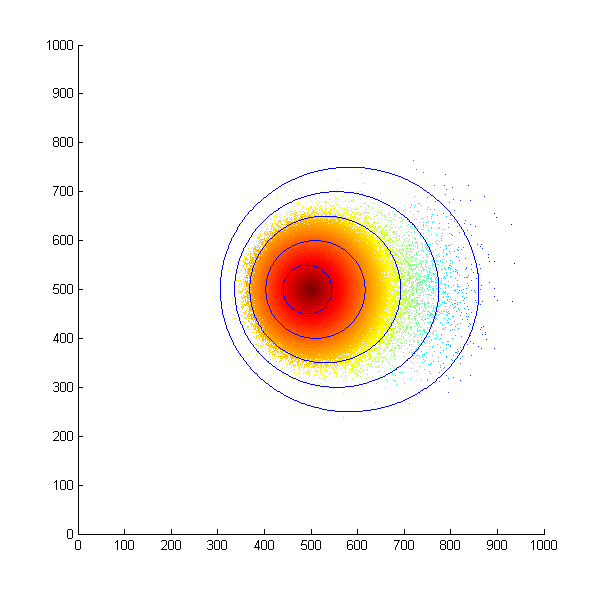
With this assumption in mind, we first simulated the situation that all the bacteria start at the same position and move in a concentration gradient. Every time the bacteria choose to swim, they will move a unit length. The following is the scatter plot after 500 steps.
Simulated E. Coli movement. All the points started from (500, 500) and moved according to the model we proposed. The color from red to blue indicates the distance the bacteria migrated from the starting point. The blue eclipse shows the approximate shape of the colony formed.
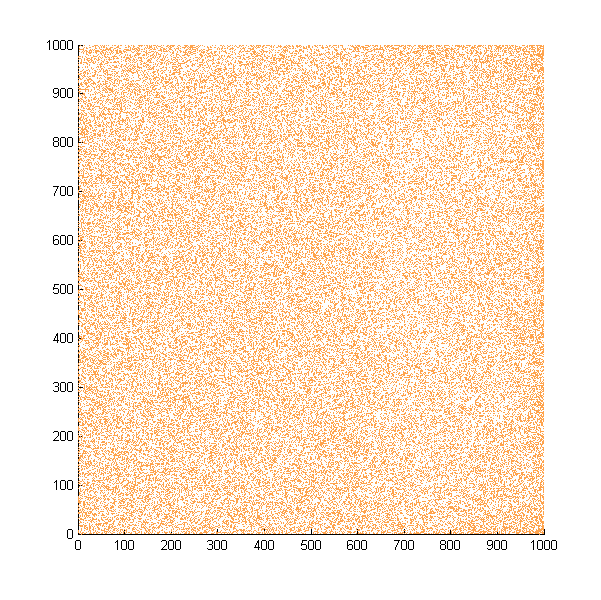
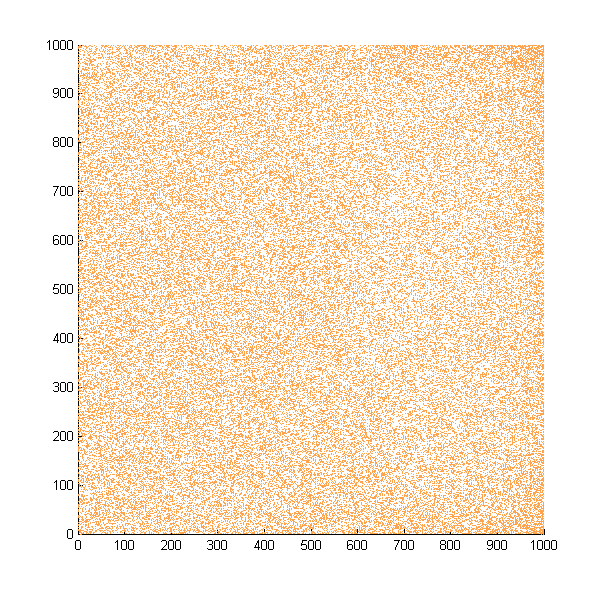
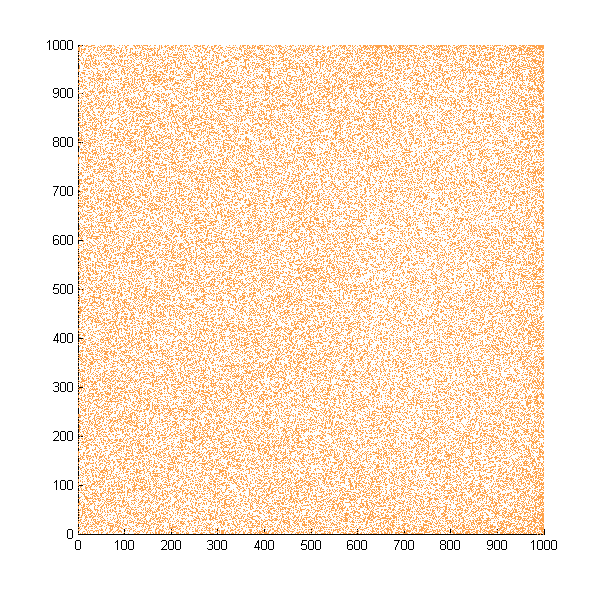
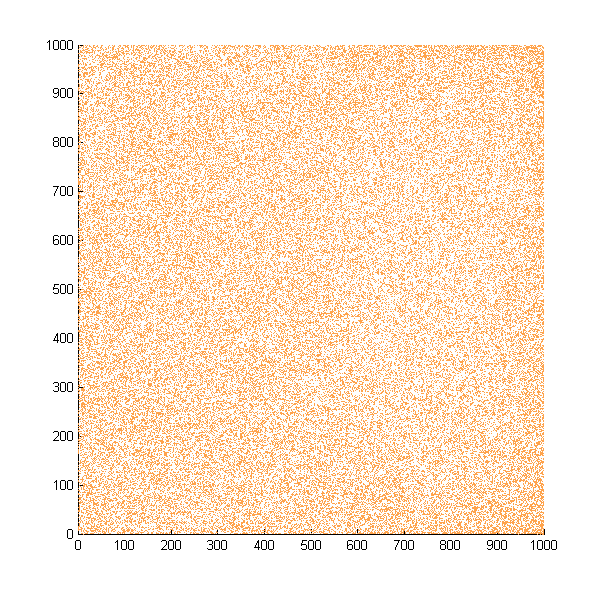
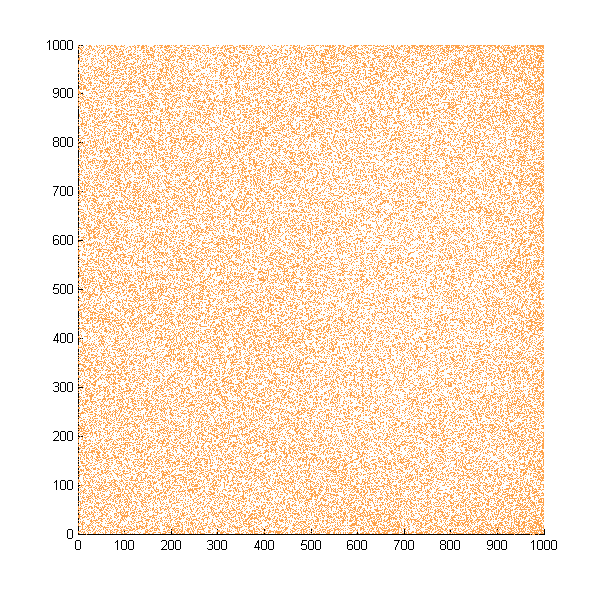
Then we move onto the pattern formed by bacteria in a gradient in liquid. We assume that bacteria was uniformed in the media before the formation of the gradient and plotted the movement after 500, 1000, 1500, 2000, and 2500 steps.
Simulated E. coli movement in liquid. The bacteria were uniformly distributed in the liquid at first and the program simulated the migration in a gradient. From x=0 to x=1000, the concentration of the chemoattractant increases, driving bacteria towards the right.
The region for E. coli to move is from x=0, y=0 to x=1000, y=1000. When a bacterium collides with the boundary, it will choose a new direction randomly. A counter was set to count all the bacteria which have crossed x=1000 at least once.
No obvious movement is seen in our simulation, but the counter reported the number of E. Coli that reached the end increases steadily.
| 15671|- | 31227|- | 46768|- | 62123|- | 77577|}
When a bacterium reaches the end, it will randomly turn around. As it will rarely change its direction in the media rich in chemoattractant, it will migrate almost to the midpoint until it makes another turn. Hence, only a small increase in the concentration of bacteria can be seen in this simulation. Our experimental results verified this point, as no obvious enrichment of bacteria can be seen in the media. However, when we measure OD600 of the two ends of our tubes, we did observe a significant difference.
|
 "
"