Team:SJTU-BioX-Shanghai/Project/Subproject2
From 2011.igem.org
(→Results) |
(→Design) |
||
| Line 66: | Line 66: | ||
The function of Switch is characterized by the amount of luciferase expressed. | The function of Switch is characterized by the amount of luciferase expressed. | ||
| - | The amount of luciferase expressed is reflected by the light emitted when luciferase acts on the appropriate luciferin substrate | + | The amount of luciferase expressed is reflected by the light emitted when luciferase acts on the appropriate luciferin substrate. |
The light can be measured by luminometer and the quantity is positively correlated with the amount of luciferase and its activity ([[Team:SJTU-BioX-Shanghai/Notebook/Protocol#Luciferase_Assay|learn more...]]). | The light can be measured by luminometer and the quantity is positively correlated with the amount of luciferase and its activity ([[Team:SJTU-BioX-Shanghai/Notebook/Protocol#Luciferase_Assay|learn more...]]). | ||
Revision as of 20:00, 5 October 2011
|
|
Stop-Codon SwitchBackgroundThe Rare Codon Modulator can only regulate the amount of target protein. To make our device a strict molecular switch that turns on/off protein biosynthesis, we need to eliminate the background. In recent years, stop codons (codonST) and stop codon suppressor tRNAs (tRNASS) are used to incorporate unnatural amino acids in protein biosynthesis. Peter Schultz and his co-workers have made a lot of contributions in this field. The advantage of codonST and tRNASS is that they can incorporate target amino acids without background noise. We make use of codonST and tRNASS as the controlling element for protein biosynthesis. IntroductionWe design a Stop-Codon Switch to turn on/off protein translation. We can control the translation process by controlling whether the ribosome can get through the stop codon placed in the target protein's mRNA. This process can be achieved by using Modulator: control the existence of charged tRNASS that recognizes the stop codon. This process is controlled by two elements:
Reporter: a stop codon is put immediately after the initial codon ATG in the target protein’s mRNA. DesignModulator:
Reporter:
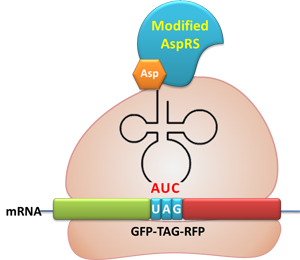
GFP and RFP linked with a flexible chain. A stop codon TAG is inserted in the flexible chain. Action: If the ribosome can get through the stop codon with the help of Modulator, both GFP and RFP can be expressed. If the ribosome cannot get through the stop codon, only GFP will be expressed.
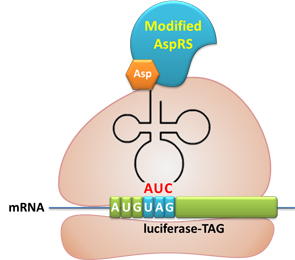
We use luciferase as our reporter gene for quantitative analysis. A TAG codon is inserted after the initial codon of the gene. Action: If the ribosome can get through the stop codon with the help of Rare-Codon Switch, luciferase can be expressed. Otherwise, luciferase cannot be expressed. The function of Switch is characterized by the amount of luciferase expressed. The amount of luciferase expressed is reflected by the light emitted when luciferase acts on the appropriate luciferin substrate. The light can be measured by luminometer and the quantity is positively correlated with the amount of luciferase and its activity (learn more...). ResultsBased on the design of Stop-Codon Switch, we have tested whether the modified AspRS can charge tRNAAsp-TAG with Asp. Both qualitative and quantitative tests are carried out.
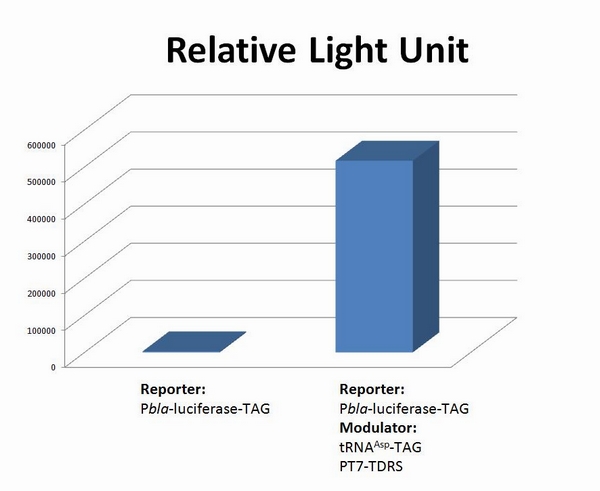 fig 1. Functional Analysis of Stop-Codon Switch. ER2566 cannot produce luciferase with Pbla-Luc-TAG (BBa_K567003) only. When BBa_K567011 PT7-TDRS and BBa_K567013 tRNAAsp-TAG are also transformed into the cell, luciferase is produced. The results proved Stop-Codon Switch as a strict molecular switch without background noise. Referance1.Cropp, T.A. and P.G. Schultz, An expanding genetic code. Trends Genet, 2004. 20(12): p. 625-30. 2.Liu, C.C., et al., Efficient expression of tyrosine-sulfated proteins in E. coli using an expanded genetic code. Nat Protoc, 2009. 4(12): p. 1784-9. 3.Liu, C.C. and P.G. Schultz, Adding new chemistries to the genetic code. Annu Rev Biochem. 79: p. 413-44. 4.Liu, D.R., T.J. Magliery, and P.G. Schultz, Characterization of an 'orthogonal' suppressor tRNA derived from E. coli tRNA2(Gln). Chem Biol, 1997. 4(9): p. 685-91. 5.Ryu, Y. and P.G. Schultz, Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat Methods, 2006. 3(4): p. 263-5. 6.Wang, F., et al., Genetic incorporation of unnatural amino acids into proteins in Mycobacterium tuberculosis. PLoS One. 5(2): p. e9354. 7.Wang, L., et al., Expanding the genetic code of Escherichia coli. Science, 2001. 292(5516): p. 498-500. 8.Wang, L. and P.G. Schultz, A general approach for the generation of orthogonal tRNAs. Chem Biol, 2001. 8(9): p. 883-90. 9.Wang, L. and P.G. Schultz, Expanding the genetic code. Chem Commun (Camb), 2002(1): p. 1-11. 10.Wang, L., J. Xie, and P.G. Schultz, Expanding the genetic code. Annu Rev Biophys Biomol Struct, 2006. 35: p. 225-49. 11.Xie, J. and P.G. Schultz, Adding amino acids to the genetic repertoire. Curr Opin Chem Biol, 2005. 9(6): p. 548-54. 12.Xie, J. and P.G. Schultz, An expanding genetic code. Methods, 2005. 36(3): p. 227-38. 13.Young, T.S. and P.G. Schultz, Beyond the canonical 20 amino acids: expanding the genetic lexicon. J Biol Chem. 285(15): p. 11039-44. 14.Zhang, Y., et al., Crystal structures of apo wild-type M. jannaschii tyrosyl-tRNA synthetase (TyrRS) and an engineered TyrRS specific for O-methyl-L-tyrosine. Protein Sci, 2005. 14(5): p. 1340-9.
|
 "
"