Team:SJTU-BioX-Shanghai/Project/Subproject1-3
From 2011.igem.org
(→Location of Rare Codons) |
(→Number of Rare Codons) |
||
| Line 11: | Line 11: | ||
==Number of Rare Codons== | ==Number of Rare Codons== | ||
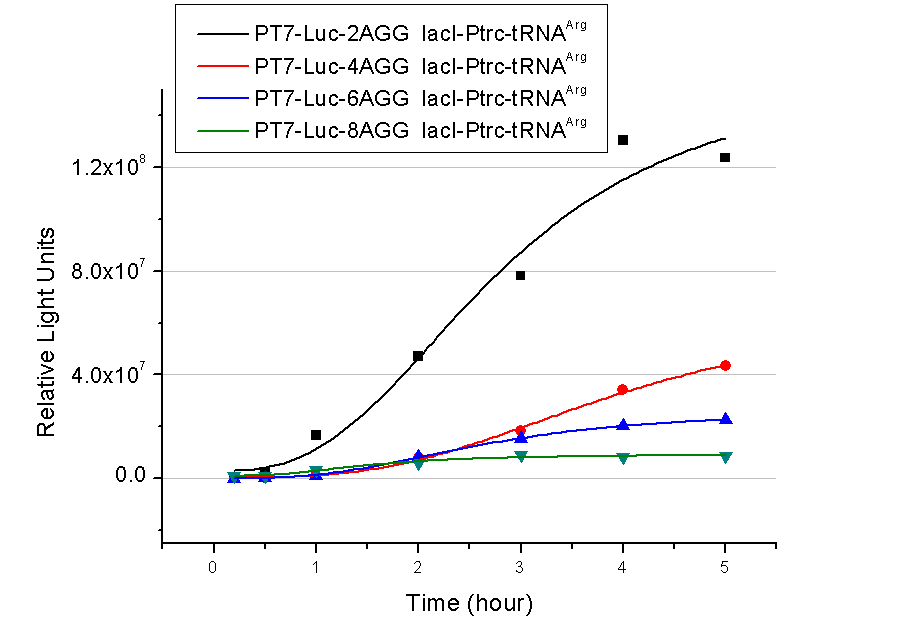
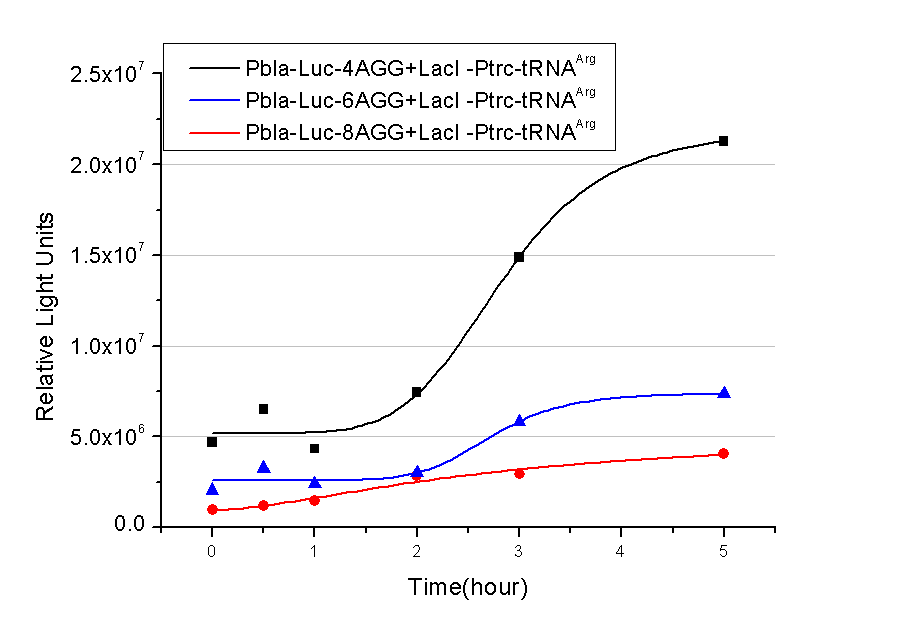
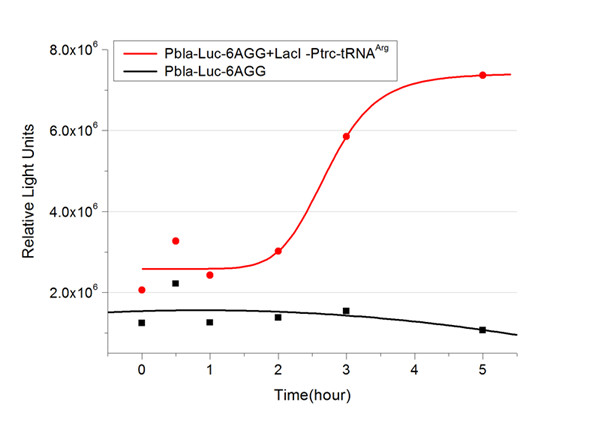
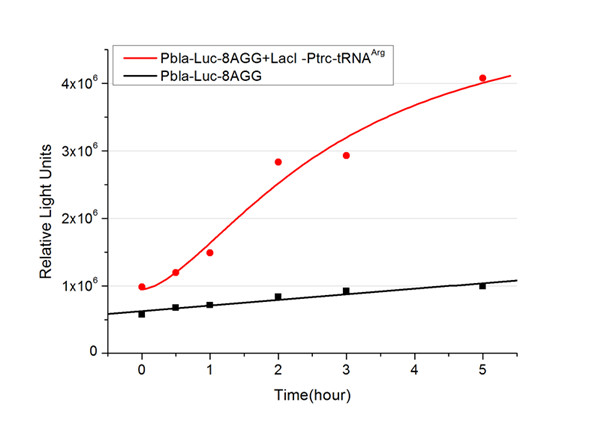
| - | In this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or bla promoter[1] are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation[1]. | + | In this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or ''bla'' promoter[1] are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation[1]. |
| - | 1) bla promoter-luciferase (weaker promoter) | + | 1) ''bla'' promoter-luciferase (weaker promoter) |
A tandem of 2, 4, 6 or 8 AGG codons is inserted after the ATG codon of wild type luciferase | A tandem of 2, 4, 6 or 8 AGG codons is inserted after the ATG codon of wild type luciferase | ||
| Line 34: | Line 34: | ||
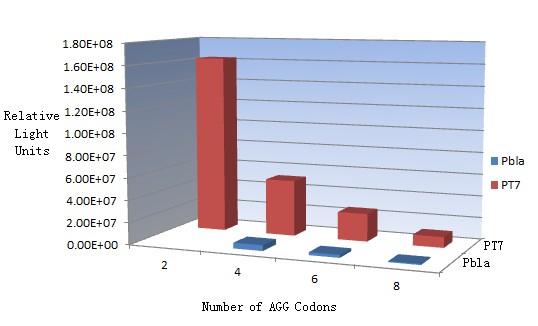
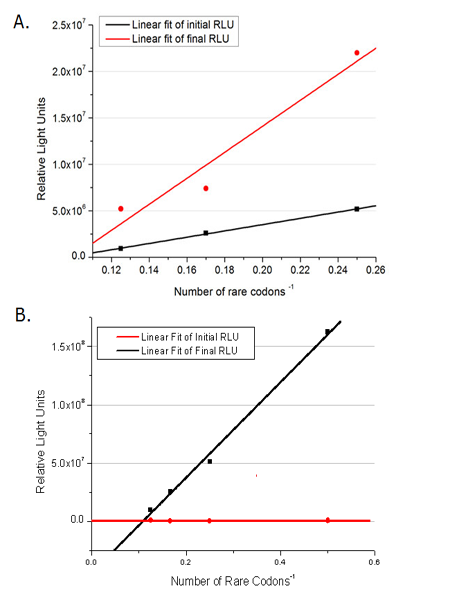
This picture reflects more clearly that the more rare codons are inserted, the lower the background expression and the narrower the range of device regulation. We are able to predict the outcome of influence of different number of rare codons in protein biosynthesis, offering valuable information for device usage. | This picture reflects more clearly that the more rare codons are inserted, the lower the background expression and the narrower the range of device regulation. We are able to predict the outcome of influence of different number of rare codons in protein biosynthesis, offering valuable information for device usage. | ||
| - | [[image:11sjtu_Compare_DATA_Fitting.png|frame|center|''Fig.11'' The comparison between background expression and induced expression of luciferase with different rare codon insertions. (A)PT7-luc reporters. (B) | + | [[image:11sjtu_Compare_DATA_Fitting.png|frame|center|''Fig.11'' The comparison between background expression and induced expression of luciferase with different rare codon insertions. (A)PT7-luc reporters. (B) P''bla''-luc reporters]] |
===Influence of different strengths of target protein promoters=== | ===Influence of different strengths of target protein promoters=== | ||
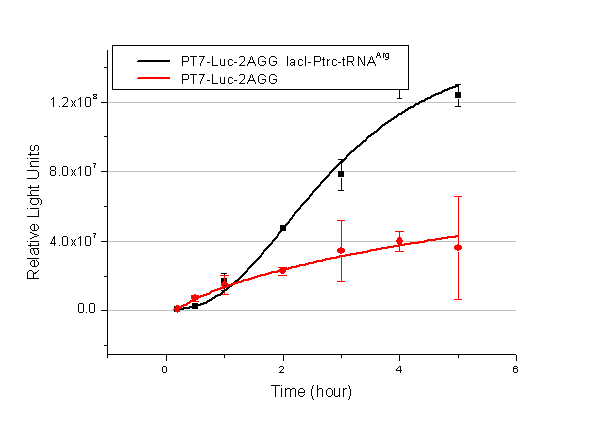
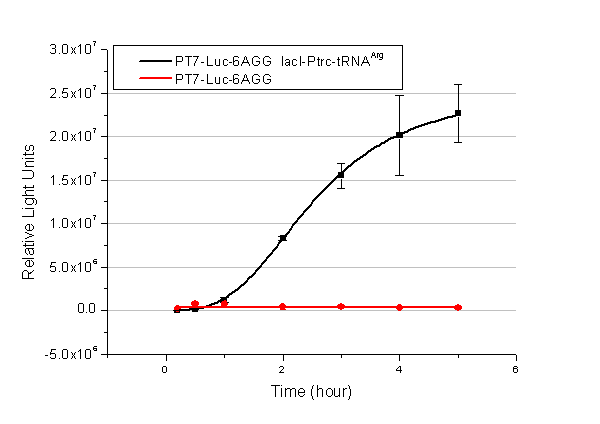
| - | We examined the influence of different Reporter promoters on the working curve of our device, which is reflected by luciferase activity. The working range of our device is pre-defined by the strength of target protein promoter, T7 promoter and bla promoter in our project. | + | We examined the influence of different Reporter promoters on the working curve of our device, which is reflected by luciferase activity. The working range of our device is pre-defined by the strength of target protein promoter, T7 promoter and ''bla'' promoter in our project. |
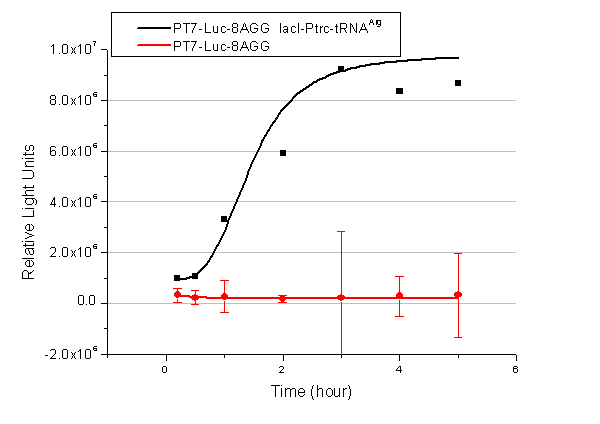
[[image:11sjtu_Compare_8.png|frame|center|''Fig.8'' (A) Working curve of tRNA Modulator ''lacI''-Ptrc-tRNA<sup>Arg</sup> under Reporter P''bla''-Luc-8AGG reflected by bioluminescence emitted from the luciferin reaction. (B) Working curve of tRNA Modulator under Reporter PT7-Luc-8AGG. Here we analyze the influences of strong/weak promoter in the working curve of tRNA Modulator. | [[image:11sjtu_Compare_8.png|frame|center|''Fig.8'' (A) Working curve of tRNA Modulator ''lacI''-Ptrc-tRNA<sup>Arg</sup> under Reporter P''bla''-Luc-8AGG reflected by bioluminescence emitted from the luciferin reaction. (B) Working curve of tRNA Modulator under Reporter PT7-Luc-8AGG. Here we analyze the influences of strong/weak promoter in the working curve of tRNA Modulator. | ||
| Line 68: | Line 68: | ||
From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced lacI-Ptrc-tRNA<sup>Arg</sup> (BBa_K567001) compared with UV excitation induced sulA promoter-tRNA<sup>Arg</sup>(BBa_K567002), though sulA promoter-tRNA<sup>Arg</sup> responded quicker to signals. So in the above experiments, we test with lacI-Ptrc-tRNA<sup>Arg</sup>. | From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced lacI-Ptrc-tRNA<sup>Arg</sup> (BBa_K567001) compared with UV excitation induced sulA promoter-tRNA<sup>Arg</sup>(BBa_K567002), though sulA promoter-tRNA<sup>Arg</sup> responded quicker to signals. So in the above experiments, we test with lacI-Ptrc-tRNA<sup>Arg</sup>. | ||
| - | |||
==Location of Rare Codons== | ==Location of Rare Codons== | ||
Revision as of 16:51, 28 October 2011
|
|
Number of Rare CodonsIn this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or bla promoter[1] are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation[1]. 1) bla promoter-luciferase (weaker promoter) A tandem of 2, 4, 6 or 8 AGG codons is inserted after the ATG codon of wild type luciferase 2) T7 promoter-luciferase (stronger promoter) A tandem of 2, 4, 6 or 8 AGG codons is inserted after the ATG codon of wild type luciferase Influence of inserted AGG codon numberThe influence of different number of rare codons in regulating protein biosynthesis is shown below: Results show that the more rare codons are inserted, the lower the background expression and the narrower the range our device can regulate. This picture reflects more clearly that the more rare codons are inserted, the lower the background expression and the narrower the range of device regulation. We are able to predict the outcome of influence of different number of rare codons in protein biosynthesis, offering valuable information for device usage. Influence of different strengths of target protein promotersWe examined the influence of different Reporter promoters on the working curve of our device, which is reflected by luciferase activity. The working range of our device is pre-defined by the strength of target protein promoter, T7 promoter and bla promoter in our project. 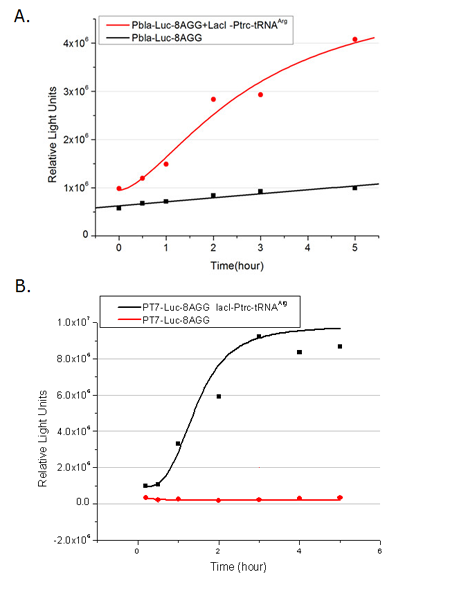 Fig.8 (A) Working curve of tRNA Modulator lacI-Ptrc-tRNAArg under Reporter Pbla-Luc-8AGG reflected by bioluminescence emitted from the luciferin reaction. (B) Working curve of tRNA Modulator under Reporter PT7-Luc-8AGG. Here we analyze the influences of strong/weak promoter in the working curve of tRNA Modulator. Moreover, strong promoter (T7) of target gene can improve the titration curve of tRNA Modulator, indicating that tRNA Modulator works better under strong target protein promoters. Note: Our device can be used as a regulating toolWe have tested luciferin reaction in cells. We examined the changes in luciferase enzyme activity over time after rare tRNA expression is induced. The amount of luciferase is reflected indirectly by the bioluminescence emitted from the luciferin reaction. Results are shown below: 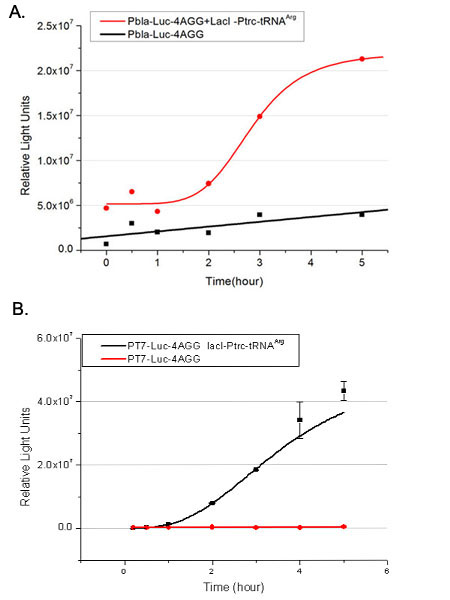 Fig.1 (A)Enzyme activity of luciferase shown by bioluminescence emitted from the luciferin reaction reflecting the working curve of tRNA Modulator lacI-Ptrc-tRNAArg (BBa_K567001) under Reporter Pbla-Luc-4AGG(BBa_K567006). (B)Enzyme activity of luciferase shown by bioluminescence emitted from the luciferin reaction reflecting the working curve of tRNA Modulator lacI-Ptrc-tRNAArg (BBa_K567001) under Reporter PT7-Luc-4AGG (BBa_K567009). The working curve of tRNA Modulator lacI-Ptrc-tRNAArg fits typical titration curve. Here we use the above two curves as examples to characterize the working curve of our device. Both curves fits typical titration curve, indicating that our device can function as a regulating tool. The rest of the working curves are shown here: Note:Click to see large figures. Results showed that all the devices’ working curves fit titration curve, indicating that our device can act as a satisfying regulating tool. From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced lacI-Ptrc-tRNAArg (BBa_K567001) compared with UV excitation induced sulA promoter-tRNAArg(BBa_K567002), though sulA promoter-tRNAArg responded quicker to signals. So in the above experiments, we test with lacI-Ptrc-tRNAArg. Location of Rare Codons |
 "
"