Team:SJTU-BioX-Shanghai/Project/Subproject1
From 2011.igem.org
(→Introduction) |
(→Rare-Codon Switch) |
||
| Line 15: | Line 15: | ||
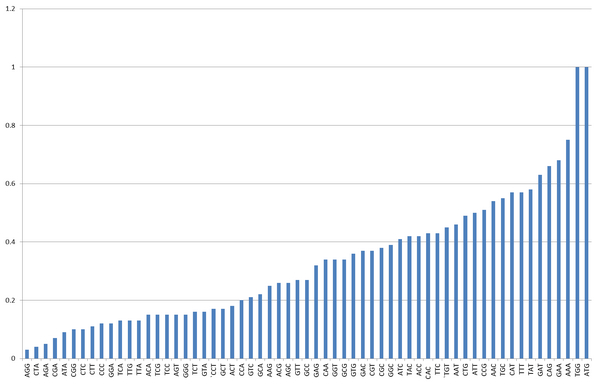
Codon degeneration indicates that more than one codon can be used to encode an amino acid. Every species has '''codon usage bias'''. For one species, some codons are rarely used(less than 1%). These codons are called rare codons. | Codon degeneration indicates that more than one codon can be used to encode an amino acid. Every species has '''codon usage bias'''. For one species, some codons are rarely used(less than 1%). These codons are called rare codons. | ||
| - | [[image:11SJTU_overview.jpg|center|frame|''Fig.1'' sense codon usage frequency in ''E.coli'']] | + | [[image:11SJTU_overview.jpg|center|frame|''Fig.1'' sense codon usage frequency in ''E.coli'' by Chunchun]] |
[[image:11SJTU_Rare_01.jpg|frame|''Fig.2'' When expressing heterogenous protein, researchers usually want to get rid of the negative effects brought by codon usage bias. | [[image:11SJTU_Rare_01.jpg|frame|''Fig.2'' When expressing heterogenous protein, researchers usually want to get rid of the negative effects brought by codon usage bias. | ||
Revision as of 03:52, 29 October 2011
|
|
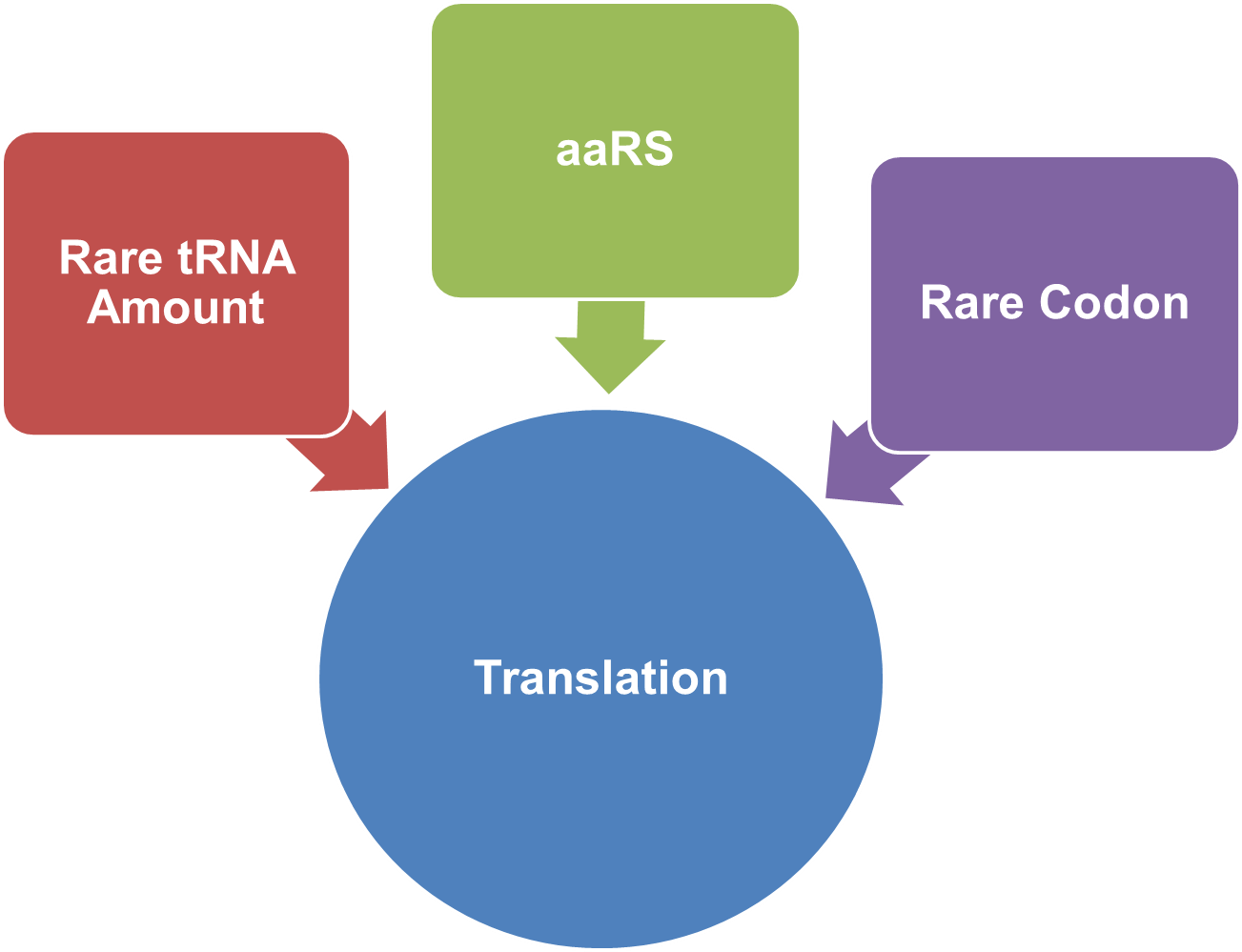
Rare-Codon SwitchIn natural biological system, translation process goes like this: aminoacyl tRNA synthetase (aaRS) charges tRNA with amino acid, and the tRNA recognizes the codon on the mRNA and brings the amino acid there. Codon degeneration indicates that more than one codon can be used to encode an amino acid. Every species has codon usage bias. For one species, some codons are rarely used(less than 1%). These codons are called rare codons. Codon usage and tRNA amount are closely related. When heterogenous protein is expressed, differences in codon usage in mRNA may impede translation because one or more tRNAs are rare in the population, especially when there are multiple consecutive rare codons at the N-terminus of a coding sequence. This has proposed a new way to control protein expression on translational level. In E.coli, the rarest used sense codon is AGG for Arg. By adding AGG codons to the site close to initial codon and regulating the abundance of rare tRNA, we can regulate protein biosynthesis on translational level.
Key WordsRare codon: codon that is used rarely or infrequently in the genome Rare tRNA: the low-abundant tRNA that can decode rare codon aaRS: aminoacyl tRNA synthetases that charges cognate tRNA with a specific amino acid
IntroductionWe design a Rare-Codon Switch controlling protein biosynthesis. We can control the translation process by controlling how well the ribosome can get through a tandem of rare codons in the target protein's mRNA. This process is controlled by two main elements: the amount of charge rare tRNA and the rare codons (including rare codon number, copy number and location) in the mRNA. Thus we can control the translation process by controlling three elements: the rare tRNA, the aaRS that charges the rare tRNA and the rare codons. See how we control the process with these three elements: Rare tRNA Amount: We over-expressed rare tRNAArg-AGG in the cell aaRS: We modified enzyme AspRS and tRNAAsp Rare Codon:We used different numbers of AGG, different locations and copies of a tandem of AGG to control protein biosynthesis |
 "
"