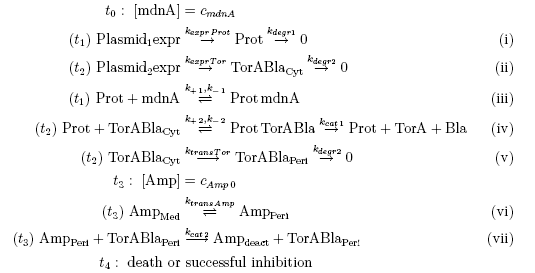Team:Potsdam Bioware/Project/Summary
From 2011.igem.org
Summary
Modification, Selection and Production of Cyclic Peptides for Therapy
One key task of biopharmaceuticals is the binding and blocking of deregulated proteins. Towards this goal, we mutate and select microviridins, which are tricyclic depsipeptides from cyanobacteria. They are small but stable due to their post-translational side-chain crosslinking. Microviridins have a high potential for therapy as they can block disease-relevant proteases. Yet, the possibilities of cyclic peptides are largely untapped since genetic systems for optimization are not well established. Thus, we developed synthetic systems for the mutation, selection and production of such peptides. We use the 6.5 kb microviridin (mdn) gene cluster cloned in E. coli plasmids, established random mutagenesis and generated focused libraries of microviridins. For selection against a panel of proteases, we are applying and testing phage display, and we are constructing a novel in-vivo selection device, which links protease blocking to antibiotic resistance. Our systems, including the 6.5 kb cluster, adhere to the BioBrick standards.
Highlights
Microviridin
The major aim of the microviridin group was to modify the mdnA such that the protease inhibiting activity is enhanced. Therefore we used random mutagenesis as well as focused oligonucleotids for creating a library, which is ready for being screened for mdnA with a therapeutically promising set of mutations. For further experiments we also fused the mdnA to a myc-tag. So in the future we will be able to purify and isolate the mdnA.
Due to the applicability of the whole mdn-cluster the creation of several BioBricks was possible. The construction was done using a given template vector containing the mdn-genes and sophisticated design of primers. Characterization of the BioBricks was done via HPLC analysis, mass spectrometry and western blot.
In a subproject we also tried to build auxiliary expression backbones with inducible promoters for easy cloning via the iGEM restriction enzyme sites. We already have the construct but the process of induction needs to be improved.[more]
Phage Display
Phage Display is an efficient tool for selecting protein or peptides with specific binding properties from a large recombinant library. This proteins are represented on the surface of bacteriophages. This enables the coupling of phenotype and stable packaged genotype because the proteins which form the phage including the proteins of interest are coded in its genome. To identify MdnA-varieties which act as protease inhibitors, a mdnA-library containing randomly mutated MdnA was created. For this purpose the phagemid vector pPARW089 was produced. This vector contains a plasmid origin of replication, so they can be amplified like plasmids additionally it contains a f1 ori which enables the packaging of single strand DNA into phages. The vector also contains the whole mdn-cluster which is needed to produce the MdnA peptide. Cloning of the mutated mdnA genes into the phagemid generates a mdnA-gene III-fusion gene. Between mdnA and gene III a myc-tag for detection is located. The successful expression of the mdnA-myc-geneIII fusion protein on the surface of the phage was determined by ELISA test using anti-myc-antibodies after transforming E. coli cells and purifying the produced phages. The next step was the performance of a phage display. To test the fundamental suitability of this screening method, phages representing mdnA on their surface and phages not representing mdnA in a ratio of one to one were incubated with immobilised trypsin which is known as a target of mdnA. After one panning round a marked concentrating of phages carrying mdnA was recognized. [more]
In Vivo Selection
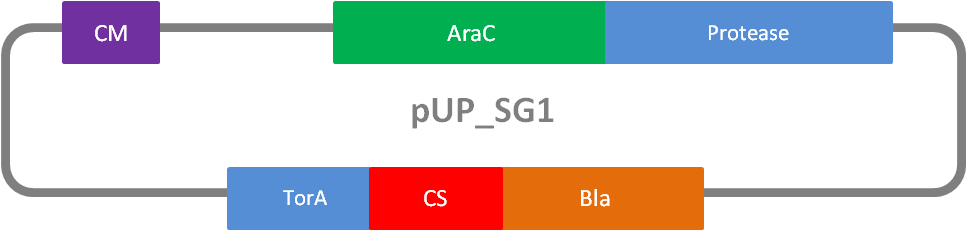
In addition to the Phage Display we developed a novel selection system. The design aimed for a cheap and time-saving alternative in contrast to an in vitro screen of protease inhibition kinetics. The assay allows us to select effective inhibitors for a any protease, among the billions of randomly generated mutants of the Microviridin. For this purpose we designed a plasmid containing two devices, first a protease activity detector and second a protease generator. We used the BioBrick <partinfo>I757010</partinfo> (β-lactamase) and <partinfo>K208005</partinfo> (ssTorA) and fused them together via a linker peptide to create our first device. The protease generator is an Arabinose inducible protease in iGEM standard.
The system works in a combined manner of the two devices. In order to confer β-lactam antibiotic resistance to cells the β-lactamase has to be exported in the periplasm. This transport is mediated by the TorA export sequence via the Twin-Arginine Translocation (TAT) system (DeLisa, 2008). The linker peptide between the TorA export sequence and the β-lactamase displays the corresponding protease cleavage site. In addition the linker peptide is chosen as short as possible to imitate folding because the TAT pathway only allows transport of correctly folded substrates. The linker peptide as well as the protease are designed as exchangeable parts.
If the protease device is functional, it will cleave the linker peptide between TorA and β-lactamase construct which leaves the cell without any antibiotic resistance. The construct was tested with increasing Ampicillin concentrations. The number of surviving colonies was depended on the export rate of the β-lactamase into the periplasm. A high cleavage rate of the linker peptide leads to a reduced Ampicillin resistance. With expressed protease a dramatically drop of the number of surviving colonies could be observed.
The survival assay was carried out with and without expressed protease. By increasing of the Ampicillin concentration we could detect a cutoff Ampicillin concentration. The colony forming units (CFU) were counted and compared.
Bilder über survival screen
We could show that our system works. The next transformation of the constructed microviridin library and our vector into competent E. coli.
Modeling
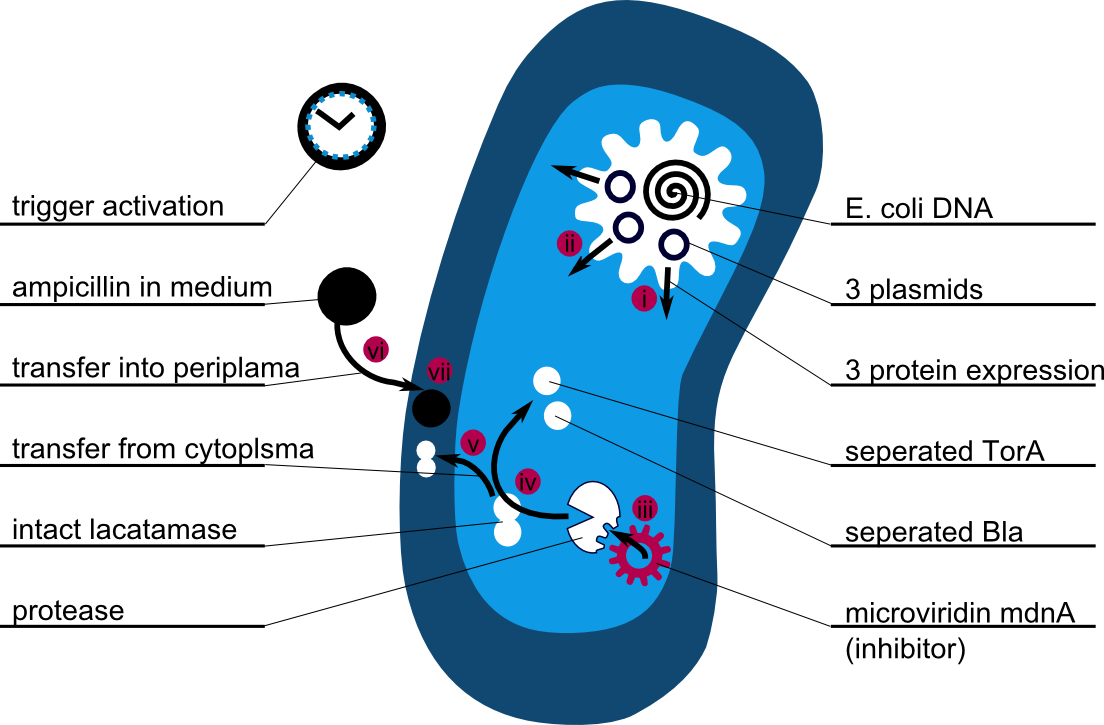
There is no synthetic biology without modeling, of course. We focused on systems modeling in which the reaction kinetics of the whole system is analysed and outcomes are predicted. Thus a synthetic biology approach can be chosen because a better understanding of the system is achieved and further changes can be planed - just like in engineering.
The following schema shows the major reactions taking place in our cell system. The Romanic numbers indicate the place of reactions that were written down as equations and then numerically propagated through time. We were able to see that our system works very well in theory. We learned about correct time-scales for our triggering and we were able to identify expected cell-division rates as a reference for the lab work.
 "
"