Team:Potsdam Bioware/Project/Details Selection
From 2011.igem.org
(→Twin Arginine Translocon (TAT)) |
(→In Vivo Selection) |
||
| Line 2: | Line 2: | ||
==In Vivo Selection== | ==In Vivo Selection== | ||
| + | ===Introduction=== | ||
| + | |||
===Background=== | ===Background=== | ||
Revision as of 01:40, 22 September 2011
Contents |
In Vivo Selection
Introduction
Background
Twin Arginine Translocon (TAT)
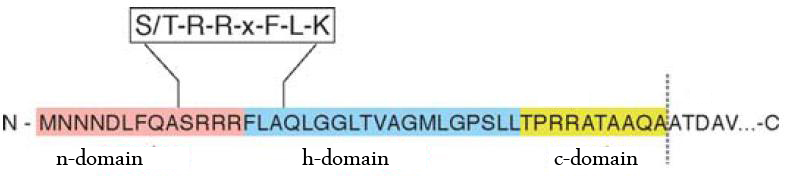
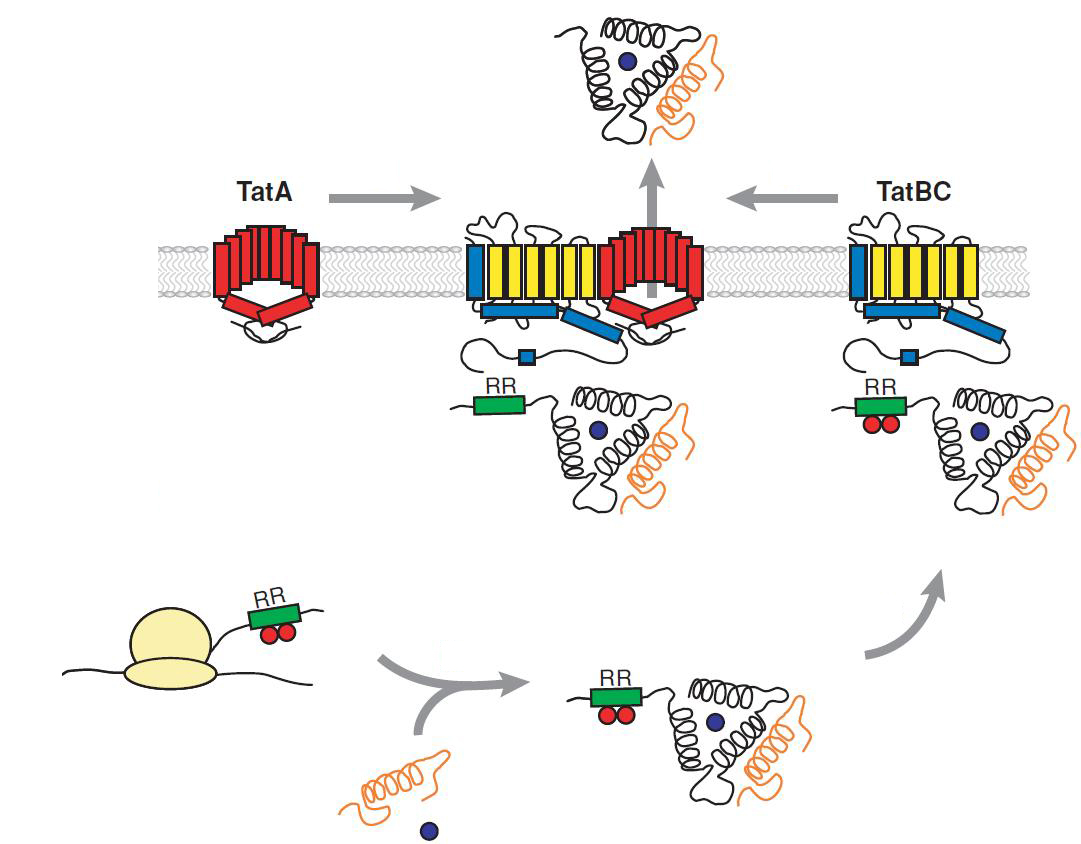
The TAT pathway is responsible for the transport of folded proteins across energy-transducing membranes and it is able of discriminating of unfolded proteins. This transport pathway is common for bacteria, archea and plants and translocates circa 6% of E.coli produced secreted proteins. The translocated proteins must have a signal peptide for targeting of the TAT-transporter, such proteins are e.g. hydrogenases, dehydrogenases and reductases. The signal sequence itself is composed of an N-terminal positive charged domain, a hydrophobic domain and a C-terminal domain.
The translocon is composed of three parts: TatA, TatB and TatC. The transport-pore is proposed to be formed during substrate binding by these three parts. The TatA, TatB and TatC proteins may form complexes of different sizes, which on the other hand form pores matching the size of the folded substrate. The transport of proteins through the TAT pathway depends on the proton motive force. Calculations by Adler & Theg showed that the transport of one folded substrate molecule requires the release of approximately 7.9x104 protons, which equals 10.000 ATP molecules. The signal sequence is cleaved off after translocation.
The used signal sequence originates from the TorA protein (Trimethylamin-N-Oxid-Reductase). It is the main respiratory enzyme which reduces TMAO under anaerobic conditions in the periplasm, where it is transported by the TAT pathway.
Reference: Philip A. Lee, Danielle Tullman-Ercekand George Georgiou Annu. Rev. Microbiol. 2006. 60:373–95
Reference: Olivier Genest, Marianne Ilbert, Vincent Méjean and Chantal Iobbi-Nivol April 22, 2005 The Journal of Biological Chemistry, 280, 15644-15648.
Tobacco Etch Virus (TEV) protease
TEV protease is the common name for the 27 kDa catalytic domain of the Nuclear Inclusion a endopeptidase (NIa) encoded by the tobacco etch virus. TEV protease is a useful reagent for cleaving fusion proteins. It recognizes a linear epitope of the general form E-Xaa-Xaa-Y -Xaa-Q-(G/S), with cleavage occurring between Q and G or Q and S. In TEV protease the serine nucleophile of the conventional Ser-Asp-His triad is a cysteine instead. This probably explains why TEV protease is resistant to many commonly used protease inhibitors.
Cabrita, L. D., Gilis, D., Robertson, A. L., Dehouck, Y., Rooman, M. and Bottomley, S.
P. (2007). Enhancing the stability and solubility of TEV protease using in silico design.
Protein Sci. 16: 2360-2367
Kapust, R. B., Tözsér, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D., and
Waugh, D. S. (2001). Tobacco etch virus protease: Mechanism of autolysis and rational
design of stable mutants with wild-type catalytic proficiency. Prot. Eng. 14: 993-1000.
Lucast, L. J., Batey, R. T., and Doudna, J. A. (2001). Large-scale purification of a stable
form of recombinant tobacco etch virus protease. Biotechniques 30: 544-550.
14_3C-Protease
Selective human enterovirus and rhinovirus inhibitors: An overview of capsid-binding and protease-inhibiting molecules. Shih SR, Chen SJ, Hakimelahi GH, Liu HJ, Tseng CT, Shia KS.
The 14_3C protease originates from the human rhinovirus. Rhinoviruses are the most frequent reason for infections of the upper and lower respiratory tract, also known as the cold. Because the 3C protease of human rhinovirus is necessary for the cleavage of the polyprotein translated from viral RNA it may serve as a potential target for development of antiviral targets. The recombinant type 14_3C protease from human rhinovirus (HRV 3C) recognizes the same cleavage site as the native enzyme: LeuGluValLeuPheGln↓GlyPro. The small, 22-kDa size of the protease got its optimal activity at 4°C but is still very active at 37 °C. It is commonly used for an easy tag removal after the purification of recombinant proteins carrying his-tag. The 14_3C works with a catalytic triade, containing the amino acid residues Ser-Asp-His at its active site.
Quelle: Human rhinovirus 3C protease as a potential target for the development of antiviral agents., Wanga QM, Chen SH., Curr Protein Pept Sci. 2007 Feb;8(1):19-27.
 "
"