Team:Potsdam Bioware/Labjournal/August part 2
From 2011.igem.org
| (2 intermediate revisions not shown) | |||
| Line 87: | Line 87: | ||
<b>Materials</b><br> | <b>Materials</b><br> | ||
| - | * DNA of pSB1T3 clones and different promotors (Ara, | + | * DNA of pSB1T3 clones and different promotors (Ara, Lac, constitutive) |
* restriction enzymes: EcoRI, XbaI | * restriction enzymes: EcoRI, XbaI | ||
| Line 259: | Line 259: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Ligation of pSB1T3 backbones with Ara, | + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Ligation of pSB1T3 backbones with Ara, Lac and constitutive promotors</h3> |
<b>Investigators:</b> Nicole, Jessica, Katharina<br> | <b>Investigators:</b> Nicole, Jessica, Katharina<br> | ||
| Line 289: | Line 289: | ||
** constitutive promotor | ** constitutive promotor | ||
| - | ** | + | ** Lac-Promotor |
* water | * water | ||
| Line 339: | Line 339: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;">Site directed mutagenesis of | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;">Site directed mutagenesis of 14_3C protease to remove iGEM restriction sites from the protease and introduction of iGEM restriction sites</h3> |
<b>For better understanding of described experiment see also: [[http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx]]</b><br> | <b>For better understanding of described experiment see also: [[http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx]]</b><br> | ||
| Line 347: | Line 347: | ||
<b>Aim:</b><br> | <b>Aim:</b><br> | ||
| - | * Removal of iGEM restriction sites from | + | * Removal of iGEM restriction sites from 14_3C protease, amplifying protease fragments with iGEM restriction sites<br> |
<b>Materials:</b><br> | <b>Materials:</b><br> | ||
| - | *Plasmid: pGEX- | + | *Plasmid: pGEX-3_14_3C |
| - | *Primers: (1) | + | *Primers: (1) f_14_3C_ACCAGC, r_14_3C_iGEM_BamHI (2) r_14_3C_ACCAGC, f_14_3C_AraFusion_NgoMIV, (3) f_14_3C_tm_Xbal208_A-T, r_14_3C_tm_Xbal208_A-T (4)r_14_3C_iGEM_BamHI, f_14_3C_tm_Xbal280_A-T |
<b> Used method: </b> | <b> Used method: </b> | ||
| Line 401: | Line 401: | ||
Expected Fragments: | Expected Fragments: | ||
| - | * | + | *14_3C_mut_fragI: 153 bp |
| - | * | + | *14_3C_mut_fragII: 66 bp |
| - | * | + | *14_3C_mut_fragIII: 72 bp |
| - | * | + | *14_3C_mut_fragIV: 260 bp |
<b> Further going: </b> | <b> Further going: </b> | ||
| - | *Assembly PCR of purificated products to produce | + | *Assembly PCR of purificated products to produce NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (mutated 14_3C-Fragment) |
<br> | <br> | ||
| Line 723: | Line 723: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;">Digestion of complete mutated TEV and | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;">Digestion of complete mutated TEV and 14_3C fragments and 3x-ligation into TEV- or 14_3C-backbones with amplified and digested AraC fragment</h3> |
<b>Investigators:</b> Paul, Stefan | <b>Investigators:</b> Paul, Stefan | ||
| Line 731: | Line 731: | ||
(1)NgoMIV_iGEM_TEV-Protease_iGEM_BamHI | (1)NgoMIV_iGEM_TEV-Protease_iGEM_BamHI | ||
| - | (2) | + | (2)NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI |
(3)HindIII_iGEM_AraC_NgoMIV (digested) | (3)HindIII_iGEM_AraC_NgoMIV (digested) | ||
| Line 739: | Line 739: | ||
* resolving of digested fragments on preparative agarose gel and excission of corresponding bands. Extraction of DNA from the gel was performed using NucleoSping ExtractII KIT. | * resolving of digested fragments on preparative agarose gel and excission of corresponding bands. Extraction of DNA from the gel was performed using NucleoSping ExtractII KIT. | ||
| - | Ligation of digested proteases into digested the pJC354- | + | Ligation of digested proteases into digested the pJC354-14_3C/TEV vectors including the AraC fragment (3) |
concentrations for ligations were calculated using gibthon ligation calculator: [http://www.gibthon.org/ligate.html CALC] | concentrations for ligations were calculated using gibthon ligation calculator: [http://www.gibthon.org/ligate.html CALC] | ||
| Line 777: | Line 777: | ||
**pSB1A3-YFP +Ara (clone C) 14: 221.4 ng/µl | **pSB1A3-YFP +Ara (clone C) 14: 221.4 ng/µl | ||
| - | **pSB1A3-CFP + | + | **pSB1A3-CFP +Lac (clone A) 15: 319.3 ng/µl |
| - | **pSB1A3-CFP + | + | **pSB1A3-CFP +Lac (clone B) 34: 105.5 ng/µl |
| - | **pSB1A3-CFP + | + | **pSB1A3-CFP +Lac (clone C) 33: 135.7 ng/µl |
| - | **pSB1A3-YFP + | + | **pSB1A3-YFP +Lac (clone A) 16: 110.6 ng/µl |
| - | **pSB1A3-YFP + | + | **pSB1A3-YFP +Lac (clone B) 4: 340.4 ng/µl |
| - | **pSB1A3-YFP + | + | **pSB1A3-YFP +Lac (clone C) 13: 290.6 ng/µl |
**pSB1A3-CFP + const (clone A) 11: 312.2 ng/µl | **pSB1A3-CFP + const (clone A) 11: 312.2 ng/µl | ||
| Line 809: | Line 809: | ||
**pSB1K3-YFP +Ara (clone C) 30: 135.1 ng/µl | **pSB1K3-YFP +Ara (clone C) 30: 135.1 ng/µl | ||
| - | **pSB1K3-CFP + | + | **pSB1K3-CFP +Lac (clone A) 20: 138.4 ng/µl |
| - | **pSB1K3-CFP + | + | **pSB1K3-CFP +Lac (clone B) 21: 58.3 ng/µl |
| - | **pSB1K3-CFP + | + | **pSB1K3-CFP +Lac (clone C) 23: 122.0 ng/µl |
| - | **pSB1K3-YFP + | + | **pSB1K3-YFP +Lac (clone A) 29: 69.3 ng/µl |
| - | **pSB1K3-YFP + | + | **pSB1K3-YFP +Lac (clone B) 17: 153.1 ng/µl |
| - | **pSB1K3-YFP + | + | **pSB1K3-YFP +Lac (clone C) 18: 90.9 ng/µl |
**pSB1K3-CFP + const (clone A) 26: 89.8 ng/µl | **pSB1K3-CFP + const (clone A) 26: 89.8 ng/µl | ||
| Line 867: | Line 867: | ||
<b>Further tasks:</b><br> | <b>Further tasks:</b><br> | ||
| - | Ligation with TEV and | + | Ligation with TEV and 14_3C and appropriate backbone<br> |
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Ligation and transformation of | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Ligation and transformation of 14_3C with backbone</h3> |
<b>Investigator:</b> Paul, Stefan<br> | <b>Investigator:</b> Paul, Stefan<br> | ||
| Line 875: | Line 875: | ||
<b>Aim:</b><br> | <b>Aim:</b><br> | ||
| - | *liagte and transform | + | *liagte and transform 14_3C with AraC and backbone<br> |
<b>Materials:</b><br> | <b>Materials:</b><br> | ||
| Line 1,135: | Line 1,135: | ||
<h2 style="background-color: rgb(240, 20, 70);">71th Labday 2011-08-21</h2> | <h2 style="background-color: rgb(240, 20, 70);">71th Labday 2011-08-21</h2> | ||
| - | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Transformation of competent RV cells with ligation products of pSB1T3/pSB1C3 backbones with Ara, | + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Transformation of competent RV cells with ligation products of pSB1T3/pSB1C3 backbones with Ara, Lac and constitutive promotors</h3> |
<b>Investigator:</b> Katharina<br> | <b>Investigator:</b> Katharina<br> | ||
| Line 1,211: | Line 1,211: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Digest of pJC354_ssTorA_NheI_CS- | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Digest of pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL</h3> |
<b>Investigator:</b> Sascha, Sebastian<br> | <b>Investigator:</b> Sascha, Sebastian<br> | ||
| Line 1,217: | Line 1,217: | ||
<b>Aim:</b><br> | <b>Aim:</b><br> | ||
| - | *Digestion of vector pJC354_ssTorA_NheI_CS- | + | *Digestion of vector pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL for ligation with AraC and 14_3C protease<br> |
<b>Materials:</b><br> | <b>Materials:</b><br> | ||
| - | * 4 µl of 3 different fractions of pJC354_ssTorA_NheI_CS- | + | * 4 µl of 3 different fractions of pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL (approx. 1,2-1,5 µg DNA)<br> |
* Restriction enzymes BamHI HF and HindIII (purchased form NEB)<br> | * Restriction enzymes BamHI HF and HindIII (purchased form NEB)<br> | ||
| Line 1,233: | Line 1,233: | ||
<b>Results:</b><br> | <b>Results:</b><br> | ||
| - | * 3 different digested vector fractions of pJC354_ssTorA_NheI_CS- | + | * 3 different digested vector fractions of pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL for ligation with AraC and 14_3C protease<br> |
<b>Further tasks:</b><br> | <b>Further tasks:</b><br> | ||
| Line 1,241: | Line 1,241: | ||
<b>Output:</b><br> | <b>Output:</b><br> | ||
| - | * pJC354_ssTorA_NheI_CS- | + | * pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL vector fraction 1, c= ng/ml<br> |
| - | * pJC354_ssTorA_NheI_CS- | + | * pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL vector fraction 2, c= ng/ml<br> |
| - | * pJC354_ssTorA_NheI_CS- | + | * pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL vector fraction 3, c= ng/ml<br> |
<h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Assembly PCR of different TEV mutagenesis fraction </h3> | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Assembly PCR of different TEV mutagenesis fraction </h3> | ||
| Line 1,395: | Line 1,395: | ||
<b>idea:</b> | <b>idea:</b> | ||
| - | [[File: UP_pUP089_plan | + | [[File: UP_pUP089_plan.jpg|350px]]<br/> |
* digest w/ AvaII and religation | * digest w/ AvaII and religation | ||
| Line 1,975: | Line 1,975: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Digest of amplified | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Digest of amplified 14_3C protease (assembled fragments after sidedirected mutagenesis), amplified AraC (from pBAD_iGEM_express mVenus), plasmid pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL and plasmid pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL</h3> |
<b>Investigator: </b> Sebastian, Sascha<br> | <b>Investigator: </b> Sebastian, Sascha<br> | ||
| Line 1,993: | Line 1,993: | ||
* PCR amplified AraC-fragment<br> | * PCR amplified AraC-fragment<br> | ||
| - | * PCR amplified and mutated | + | * PCR amplified and mutated 14_3C protease<br> |
<b>Methode:</b><br> | <b>Methode:</b><br> | ||
| Line 2,001: | Line 2,001: | ||
<i>Reaction batches:</i><br> | <i>Reaction batches:</i><br> | ||
| - | * Batch 1: 21.7 µl | + | * Batch 1: 21.7 µl 14_3C fraction I (c=157.0 ng/µl), 2.5 µl NgoMIV, 2.5 µl BamHI, 0.3 µl BSA, 3 µl Buffer 4<br> |
| - | * Batch 2: 21.7 µl | + | * Batch 2: 21.7 µl 14_3C fraction II(c=157.5 ng/µl), 2.5 µl NgoMIV, 2.5 µl BamHI, 0.3 µl BSA, 3 µl Buffer 4<br> |
* Batch 3: 22.0 µl AraC (c=175.0 ng/µl), 2.5 µl NgoMIV, 2.5 µl HindIII, 3 µl Buffer 2<br> | * Batch 3: 22.0 µl AraC (c=175.0 ng/µl), 2.5 µl NgoMIV, 2.5 µl HindIII, 3 µl Buffer 2<br> | ||
| Line 2,023: | Line 2,023: | ||
<b>Output:</b><br> | <b>Output:</b><br> | ||
| - | * digested fragments of AraC, | + | * digested fragments of AraC, 14_3C protease and plasmid backbones for transformation (AraC + 14_3C + "14_3C-backbone", AraC + TEV + "TEV-backbone")<br> |
1) EcoRI_AraC_NgoMIV<br> | 1) EcoRI_AraC_NgoMIV<br> | ||
| Line 2,029: | Line 2,029: | ||
2) HindIII_AraC_NgoMIV<br> | 2) HindIII_AraC_NgoMIV<br> | ||
| - | 3) | + | 3) NgoMIV_14_3C_BamHI Batch 1<br> |
| - | 4) | + | 4) NgoMIV_14_3C_BamHI Batch 2<br> |
5) BamHI_pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL_HindIII Batch 1<br> | 5) BamHI_pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL_HindIII Batch 1<br> | ||
| Line 2,167: | Line 2,167: | ||
**pSB1A3-YFP +Ara (clone C) 14: 221.4 ng/µl | **pSB1A3-YFP +Ara (clone C) 14: 221.4 ng/µl | ||
| - | **pSB1A3-CFP + | + | **pSB1A3-CFP +Lac (clone A) 15: 319.3 ng/µl |
| - | **pSB1A3-CFP + | + | **pSB1A3-CFP +Lac (clone B) 34: 105.5 ng/µl |
| - | **pSB1A3-CFP + | + | **pSB1A3-CFP +Lac (clone C) 33: 135.7 ng/µl |
| - | **pSB1A3-YFP + | + | **pSB1A3-YFP +Lac (clone A) 16: 110.6 ng/µl |
| - | **pSB1A3-YFP + | + | **pSB1A3-YFP +Lac (clone B) 4: 340.4 ng/µl |
| - | **pSB1A3-YFP + | + | **pSB1A3-YFP +Lac (clone C) 13: 290.6 ng/µl |
**pSB1A3-CFP + const (clone A) 11: 312.2 ng/µl | **pSB1A3-CFP + const (clone A) 11: 312.2 ng/µl | ||
| Line 2,199: | Line 2,199: | ||
**pSB1K3-YFP +Ara (clone C) 30: 135.1 ng/µl | **pSB1K3-YFP +Ara (clone C) 30: 135.1 ng/µl | ||
| - | **pSB1K3-CFP + | + | **pSB1K3-CFP +Lac (clone A) 20: 138.4 ng/µl |
| - | **pSB1K3-CFP + | + | **pSB1K3-CFP +Lac (clone B) 21: 58.3 ng/µl |
| - | **pSB1K3-CFP + | + | **pSB1K3-CFP +Lac (clone C) 23: 122.0 ng/µl |
| - | **pSB1K3-YFP + | + | **pSB1K3-YFP +Lac (clone A) 29: 69.3 ng/µl |
| - | **pSB1K3-YFP + | + | **pSB1K3-YFP +Lac (clone B) 17: 153.1 ng/µl |
| - | **pSB1K3-YFP + | + | **pSB1K3-YFP +Lac (clone C) 18: 90.9 ng/µl |
**pSB1K3-CFP + const (clone A) 26: 89.8 ng/µl | **pSB1K3-CFP + const (clone A) 26: 89.8 ng/µl | ||
| Line 2,257: | Line 2,257: | ||
** 24.2 µl water | ** 24.2 µl water | ||
| - | *Reaction mix ( | + | *Reaction mix (Lac) |
** 0.5 µl DNA | ** 0.5 µl DNA | ||
| Line 2,437: | Line 2,437: | ||
|- | |- | ||
| - | | 23 || pSB1A3-CFP + | + | | 23 || pSB1A3-CFP +Lac (clone A) || 15|| |
|- | |- | ||
| - | | 24 || pSB1A3-CFP + | + | | 24 || pSB1A3-CFP +Lac (clone B) || 15|| |
|- | |- | ||
| - | | 25 || pSB1A3-CFP + | + | | 25 || pSB1A3-CFP +Lac (clone C) || 15|| |
|- | |- | ||
| - | | 26 || pSB1A3-YFP + | + | | 26 || pSB1A3-YFP +Lac (clone A) || 15|| |
|- | |- | ||
| - | | 27 || pSB1A3-YFP + | + | | 27 || pSB1A3-YFP +Lac (clone B)|| 15|| |
|- | |- | ||
| - | | 28 || pSB1A3-YFP + | + | | 28 || pSB1A3-YFP +Lac (clone C)|| 15|| |
|- | |- | ||
| - | | 29 || pSB1K3-CFP + | + | | 29 || pSB1K3-CFP +Lac (clone A)|| 15|| |
|- | |- | ||
| - | | 30 || pSB1K3-CFP + | + | | 30 || pSB1K3-CFP +Lac (clone B) || 15|| |
|- | |- | ||
| - | | 31 || pSB1K3-CFP + | + | | 31 || pSB1K3-CFP +Lac (clone C) || 15|| |
|- | |- | ||
| - | | 32 || pSB1K3-YFP + | + | | 32 || pSB1K3-YFP +Lac (clone A) || 15|| |
|- | |- | ||
| - | | 33 || pSB1K3-YFP + | + | | 33 || pSB1K3-YFP +Lac (clone B) || 15|| |
|- | |- | ||
| - | | 34 || pSB1K3-YFP + | + | | 34 || pSB1K3-YFP +Lac (clone C)|| 15|| |
|- | |- | ||
| Line 2,577: | Line 2,577: | ||
* digested, gel purified, dephoshorylated vectors: pSB1C3, pSB1T3-YFP I clone c, pSB1T3-YFP II plate 2 clone d | * digested, gel purified, dephoshorylated vectors: pSB1C3, pSB1T3-YFP I clone c, pSB1T3-YFP II plate 2 clone d | ||
| - | * Inserts: Ara promoter, | + | * Inserts: Ara promoter, Lac promoter, constitutive promoter |
<b> Method:</b> | <b> Method:</b> | ||
| Line 2,607: | Line 2,607: | ||
** pSB1C3 + Ara | ** pSB1C3 + Ara | ||
| - | ** pSB1C3 + | + | ** pSB1C3 + Lac |
** pSB1C3 + const | ** pSB1C3 + const | ||
| Line 2,615: | Line 2,615: | ||
** pSB1T3-YFP II + Ara | ** pSB1T3-YFP II + Ara | ||
| - | ** pSB1T3-YFP II + | + | ** pSB1T3-YFP II + Lac |
** pSB1T3-YFP II + const | ** pSB1T3-YFP II + const | ||
| Line 2,623: | Line 2,623: | ||
** pSB1T3-YFP I + Ara | ** pSB1T3-YFP I + Ara | ||
| - | ** pSB1T3-YFP I + | + | ** pSB1T3-YFP I + Lac |
** pSB1T3-YFP I + const | ** pSB1T3-YFP I + const | ||
| Line 2,791: | Line 2,791: | ||
<h2 style="background-color: rgb(240, 20, 70);">74th Labday 2011-08-24</h2> | <h2 style="background-color: rgb(240, 20, 70);">74th Labday 2011-08-24</h2> | ||
| - | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Transformation w/ pUP089 A and B, BBa_K40304_Affibody_MiddleLinker+Ara/Const/ | + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Transformation w/ pUP089 A and B, BBa_K40304_Affibody_MiddleLinker+Ara/Const/Lac, pSB1T3+YFPI+Ara/Const/Lac and pSB1T3+YFPII+Ara/Const/Lac </h3> |
<b>Investigators:</b> Nicole, Nadine<br> | <b>Investigators:</b> Nicole, Nadine<br> | ||
| Line 2,801: | Line 2,801: | ||
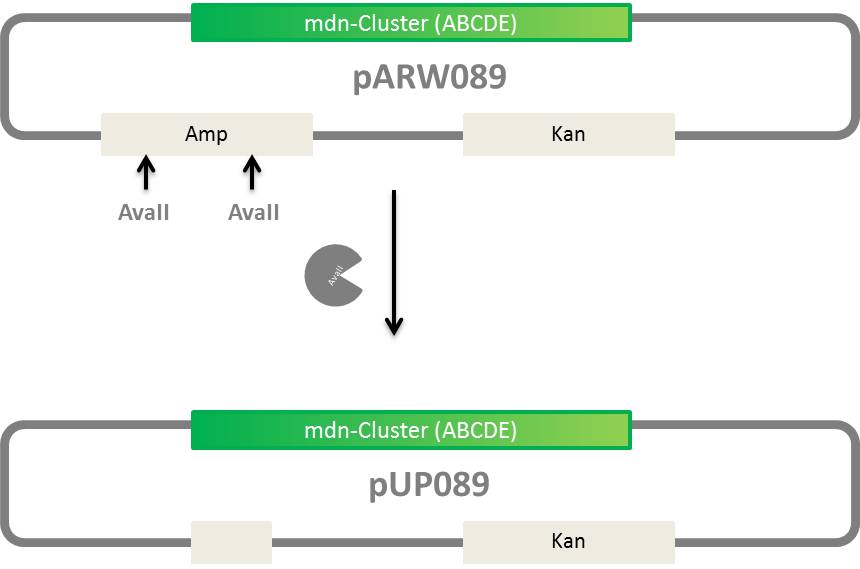
* creating pARW089 without Amp resistance, called then pUP089 | * creating pARW089 without Amp resistance, called then pUP089 | ||
| - | * get expression backbones w/ Tet or Cm resistance and Ara, Const. or | + | * get expression backbones w/ Tet or Cm resistance and Ara, Const. or Lac promotors |
<b>Material:</b><br> | <b>Material:</b><br> | ||
| Line 2,813: | Line 2,813: | ||
** pSB1C3 + Ara | ** pSB1C3 + Ara | ||
| - | ** pSB1C3 + | + | ** pSB1C3 + Lac |
** pSB1C3 + const | ** pSB1C3 + const | ||
| Line 2,821: | Line 2,821: | ||
** pSB1T3-YFP II + Ara | ** pSB1T3-YFP II + Ara | ||
| - | ** pSB1T3-YFP II + | + | ** pSB1T3-YFP II + Lac |
** pSB1T3-YFP II + const | ** pSB1T3-YFP II + const | ||
| Line 2,829: | Line 2,829: | ||
** pSB1T3-YFP I + Ara | ** pSB1T3-YFP I + Ara | ||
| - | ** pSB1T3-YFP I + | + | ** pSB1T3-YFP I + Lac |
** pSB1T3-YFP I + const | ** pSB1T3-YFP I + const | ||
| Line 3,407: | Line 3,407: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Transformation w/ pUP089 A and B, BBa_K40304_Affibody_MiddleLinker+Ara/Const/ | + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Transformation w/ pUP089 A and B, BBa_K40304_Affibody_MiddleLinker+Ara/Const/Lac, pSB1T3+YFPI+Ara/Const/Lac and pSB1T3+YFPII+Ara/Const/Lac </h3> |
<b>Investigators:</b> Nicole, Nadine<br> | <b>Investigators:</b> Nicole, Nadine<br> | ||
| Line 3,417: | Line 3,417: | ||
* creating pARW089 without Amp resistance, called then pUP089 | * creating pARW089 without Amp resistance, called then pUP089 | ||
| - | * get expression backbones w/ Tet or Cm resistance and Ara, Const. or | + | * get expression backbones w/ Tet or Cm resistance and Ara, Const. or Lac promotors |
<b>Material:</b><br> | <b>Material:</b><br> | ||
| Line 3,429: | Line 3,429: | ||
** pSB1C3 + Ara | ** pSB1C3 + Ara | ||
| - | ** pSB1C3 + | + | ** pSB1C3 + Lac |
** pSB1C3 + const | ** pSB1C3 + const | ||
| Line 3,437: | Line 3,437: | ||
** pSB1T3-YFP II + Ara | ** pSB1T3-YFP II + Ara | ||
| - | ** pSB1T3-YFP II + | + | ** pSB1T3-YFP II + Lac |
** pSB1T3-YFP II + const | ** pSB1T3-YFP II + const | ||
| Line 3,445: | Line 3,445: | ||
** pSB1T3-YFP I + Ara | ** pSB1T3-YFP I + Ara | ||
| - | ** pSB1T3-YFP I + | + | ** pSB1T3-YFP I + Lac |
** pSB1T3-YFP I + const | ** pSB1T3-YFP I + const | ||
| Line 3,551: | Line 3,551: | ||
** pSB1C3 + Ara | ** pSB1C3 + Ara | ||
| - | ** pSB1C3 + | + | ** pSB1C3 + Lac |
** pSB1C3 + const | ** pSB1C3 + const | ||
| Line 3,559: | Line 3,559: | ||
** pSB1T3-YFP II + Ara | ** pSB1T3-YFP II + Ara | ||
| - | ** pSB1T3-YFP II + | + | ** pSB1T3-YFP II + Lac |
** pSB1T3-YFP II + const | ** pSB1T3-YFP II + const | ||
| Line 3,567: | Line 3,567: | ||
** pSB1T3-YFP I + Ara | ** pSB1T3-YFP I + Ara | ||
| - | ** pSB1T3-YFP I + | + | ** pSB1T3-YFP I + Lac |
** pSB1T3-YFP I + const | ** pSB1T3-YFP I + const | ||
| Line 3,681: | Line 3,681: | ||
** pSB1C3 + Ara | ** pSB1C3 + Ara | ||
| - | ** pSB1C3 + | + | ** pSB1C3 + Lac |
** pSB1C3 + const | ** pSB1C3 + const | ||
| Line 3,689: | Line 3,689: | ||
** pSB1T3-YFP II + Ara | ** pSB1T3-YFP II + Ara | ||
| - | ** pSB1T3-YFP II + | + | ** pSB1T3-YFP II + Lac |
** pSB1T3-YFP II + const | ** pSB1T3-YFP II + const | ||
| Line 3,697: | Line 3,697: | ||
** pSB1T3-YFP I + Ara | ** pSB1T3-YFP I + Ara | ||
| - | ** pSB1T3-YFP I + | + | ** pSB1T3-YFP I + Lac |
** pSB1T3-YFP I + const | ** pSB1T3-YFP I + const | ||
| Line 3,861: | Line 3,861: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> gel purification of AraC, TEV, | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> gel purification of AraC, TEV, 14_3C and TEv vector and 14_3C vector</h3> |
<b>Investigator: </b> Stefan<br> | <b>Investigator: </b> Stefan<br> | ||
| Line 3,877: | Line 3,877: | ||
ligation of different fragmentsand transformation of competent E.coli XL1 blue cells.<br> | ligation of different fragmentsand transformation of competent E.coli XL1 blue cells.<br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> ligation of AraC, TEV/ | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> ligation of AraC, TEV/14_3C with the TEV/14_3C vector</h3> |
<b>Investigator: </b> Stefan<br> | <b>Investigator: </b> Stefan<br> | ||
| Line 3,887: | Line 3,887: | ||
|+ optional table caption | |+ optional table caption | ||
| - | ! Column heading 1 !! TEV 1 µL !! TEV 2 µL !! TEV 3 µL !! TEV 4 µL !! | + | ! Column heading 1 !! TEV 1 µL !! TEV 2 µL !! TEV 3 µL !! TEV 4 µL !! 14_3C 1 µL !! 14_3C 2 µL !! control 14_3C vector µL !! control TEV vector µL !! |
|- | |- | ||
| Line 5,918: | Line 5,918: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Production of TEV and | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Production of TEV and 14_3C biobricks part 1 </h3> |
| - | Aim: indtroduction of iGEM restriction sites to produced mutated TEV and | + | Aim: indtroduction of iGEM restriction sites to produced mutated TEV and 14_3C Fragments. |
Primer TEV: | Primer TEV: | ||
| Line 5,928: | Line 5,928: | ||
(2) r_TEV_ACCAGC, f_TEV_iGEM | (2) r_TEV_ACCAGC, f_TEV_iGEM | ||
| - | Primer | + | Primer 14_3C: |
| - | (1) | + | (1) f_14_3C_ACCAGC, r_14_3C_iGEM_Eco81l |
| - | (2) | + | (2) r_14_3C_ACCAGC, f_14_3C_iGEM |
<b>Methode:</b><br> | <b>Methode:</b><br> | ||
| Line 5,938: | Line 5,938: | ||
PCR<br> | PCR<br> | ||
| - | *Template: 1µl (TEV or | + | *Template: 1µl (TEV or 14_3C <10ng) |
*Nucleotides: 1µl of 10mM ready to use dNTP mix<br> | *Nucleotides: 1µl of 10mM ready to use dNTP mix<br> | ||
| Line 6,576: | Line 6,576: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> sequencing of TEV and | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> sequencing of TEV and 14_3C clones </h3> |
Aim: get sequences | Aim: get sequences | ||
| Line 7,298: | Line 7,298: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Assembly PCR for TEV and | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Assembly PCR for TEV and 14_3C </h3> |
| - | Aim: get the side-directed mutated TEV and | + | Aim: get the side-directed mutated TEV and 14_3C fragment<br> |
<b>Methode:</b><br> | <b>Methode:</b><br> | ||
| Line 7,310: | Line 7,310: | ||
(2) r_TEV_iGEM_BamHI<br> | (2) r_TEV_iGEM_BamHI<br> | ||
| - | Primer | + | Primer 14_3C: |
| - | (1) | + | (1) f_14_3C_iGEM<br> |
| - | (2) | + | (2) r_14_3C_iGEM_BamHI |
<b>Methode:</b><br> | <b>Methode:</b><br> | ||
| Line 7,320: | Line 7,320: | ||
PCR<br> | PCR<br> | ||
| - | *Template: 1 µL (TEV or | + | *Template: 1 µL (TEV or 14_3C <10ng) |
*Nucleotides: 1 µL of 10 mM ready to use dNTP mix<br> | *Nucleotides: 1 µL of 10 mM ready to use dNTP mix<br> | ||
| Line 7,634: | Line 7,634: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> gel electrophoresis of TEV and | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> gel electrophoresis of TEV and 14_3C </h3> |
Aim: check sizes<br> | Aim: check sizes<br> | ||
| Line 7,794: | Line 7,794: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Assembly PCR for | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Assembly PCR for 14_3C, PCR of AraC and TEV </h3> |
| - | Aim: get the side-directed mutated TEV and | + | Aim: get the side-directed mutated TEV and 14_3C fragment<br> |
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 7,806: | Line 7,806: | ||
(2) r_TEV_iGEM_BamHI<br> | (2) r_TEV_iGEM_BamHI<br> | ||
| - | Primer | + | Primer 14_3C: |
| - | (1) | + | (1) f_14_3C_iGEM<br> |
| - | (2) | + | (2) r_14_3C_iGEM_BamHI |
<b>Methode:</b><br> | <b>Methode:</b><br> | ||
| Line 7,816: | Line 7,816: | ||
PCR<br> | PCR<br> | ||
| - | *Template: 1 µL (TEV or | + | *Template: 1 µL (TEV or 14_3C <10ng) |
*Nucleotides: 1 µL of 10 mM ready to use dNTP mix<br> | *Nucleotides: 1 µL of 10 mM ready to use dNTP mix<br> | ||
Latest revision as of 13:03, 21 September 2011
66th Labday 2011-08-17
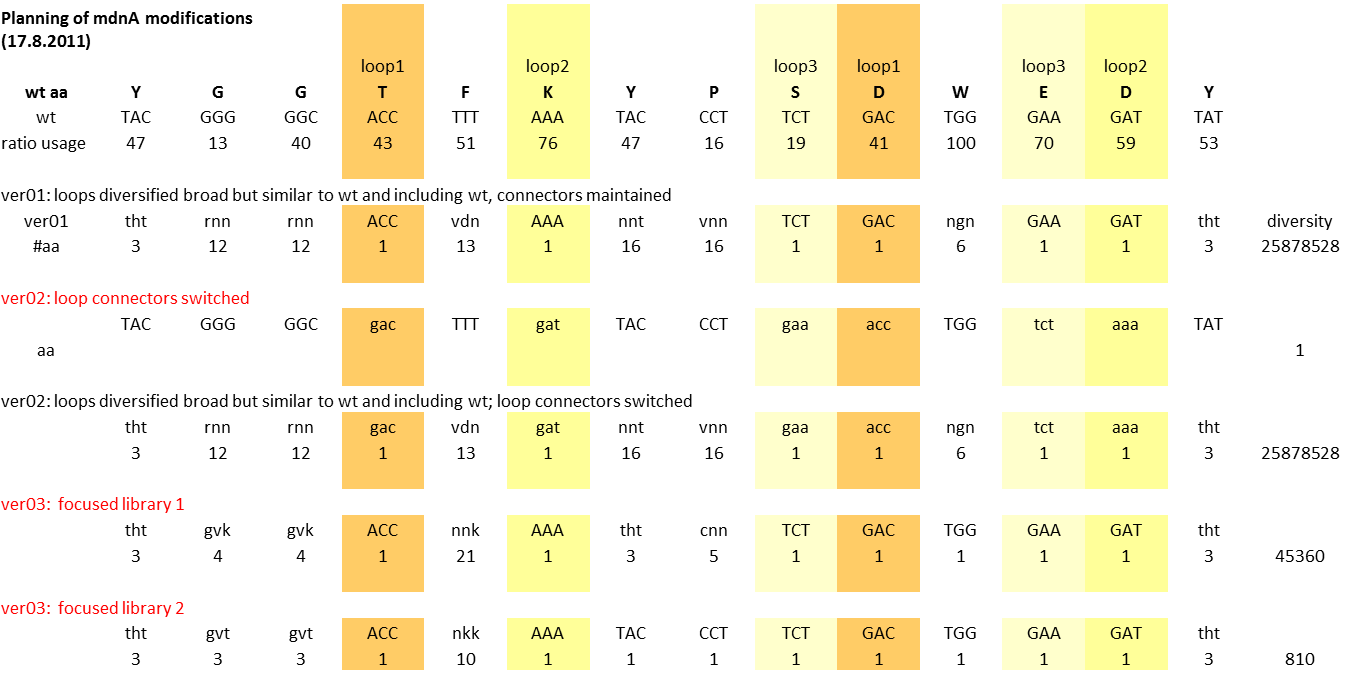
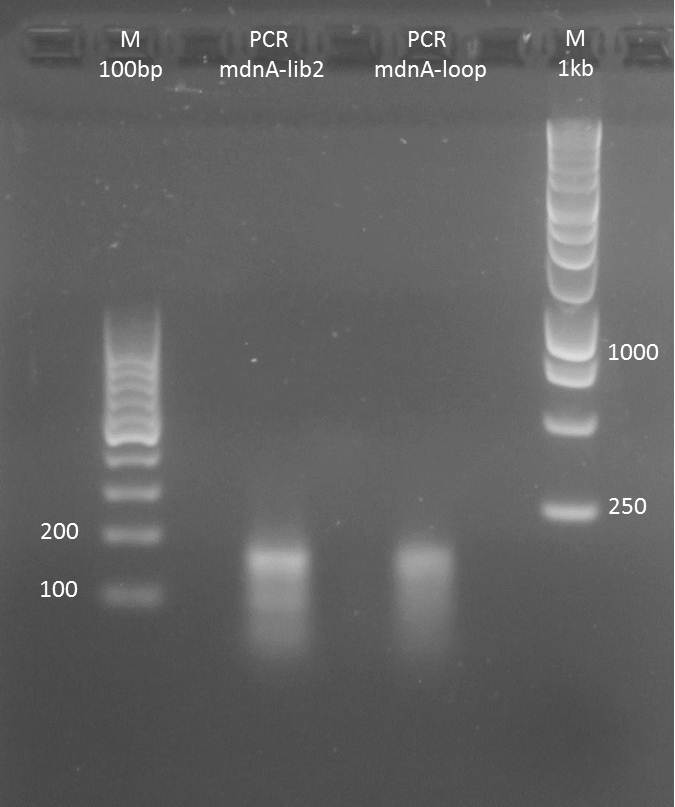
Planning mdnA modification (library)
Investigators: Nicole
Time: 2011-08-17, 20:45
Aim: Planning mdnA library
Materials
- codon table
- Kristian's diversity sheet
- excel
Results:
Output:
Excel Table: Y:\Klonierung\mdna modification\mdna modification 03.xlsx
67th Labday 2011-08-18
Ordering oligos for modified mdnA library
Investigators: Nicole
miniprep of pSB1T3 clones containing CFP and YFP, respectively
Investigators: Nicole, Jessica, Steffi, Katharina
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 30µl
- Check concentration with NanoDrop
Results:
- pSB1T3+YFP I clone b: 72.1 ng/µl
- pSB1T3+YFP I clone c: 53.5 ng/µl
- pSB1T3+YFP I clone a: 197.1 ng/µl
- pSB1T3+YFP II plate 1 clone b: 46.1 ng/µl
- pSB1T3+YFP II clone a: 62.6 ng/µl
- pSB1T3+YFP II plate 2 clone d: 25.6 ng/µl
- pSB1T3+YFP II plate 2 clone c: 25.6 ng/µl
- pSB1T3+YFP I clone d: 101.8 ng/µl
Further Tasks:
- restriction enzyme digestion
- ligation with promotors
- transformation
restriction enzyme digestion of
Investigators: Nicole, Jessica, Katharina
Materials
- DNA of pSB1T3 clones and different promotors (Ara, Lac, constitutive)
- restriction enzymes: EcoRI, XbaI
- NEB Buffer 4
- water
Method
- 0.5µl EcoRI
- 0.5µl XbaI (again 0.5µl were added after 1.5 h)
- 1000ng DNA
- water added to total volume of 30µl
- restriction enzyme digestion overnight at 37°C
Digestion of pak bla KDIR
Investigators: Sabine
Aim:
- cutting out of geneIII for using as PCR template
Reason:
- pak bla KDIR contains a part of a myc tag
- the overhang of the geneIII forward primer contains a complete myc tag for insertion of myc into the phage display vector
- this may lead to the low yield of geneIII target DNA after PCR using entire vector pak bla KDIR as template
Time: 2011-08-18,10:00-14:00
Materials/Methods:
- 5 µl pak bla KDIR (ca 4500 ng)
- 2 µl NEB 10x buffer 2
- 1 µl restriction enzyme AvaI
- 1 µ restriction enzyme HindIII
- 11 µl water
- 4 h at 37°C
Further tasks:
- gel electrophoresis for purification of geneIII
- perform PCR of geneIII
Repeated PCR of mdnA and gene III for phage display (strategy 2)
Investigator: Sabine
Time: 2011-08-18, 15:00-18:00
Aim:
- amplification of geneIII with NgoMIV and AatII and rfc 25 restriction sites (strategy 2)
Reaction Components:
- 1 µl / 6 ng DNA (cut out geneIII from pak bla KDIR)
- 0,25 µl OneTaq Polymerase
- 1 µl dNTPs
- 1 µl per primer (pf_geneIII_Xba_NgoMIV_myc and pr_geneIII_iGEM_AatII)
- 5 µl 5x PCR Buffer
- 30,75 µl DNase free water
- purification of PCR fragments with QIAquick Gel Extraction Kit (250)
Further tasks:
- digestion / ligation
Digestion of pSB1C3
Investigators: Sabine
Aim:
- cloning of biobricks mdnA, geneIII and mdna/geneIII (fusion gene) into pSB1C3
Time: 2011-08-18,17:30-18:00
Materials/Methods:
- 8 µl pSB1C3 (ca 2000 ng)
- 2 µl NEB 10x buffer 2
- 1 µl restriction enzyme XbaI
- 1 µ restriction enzyme PstI
- 0,2 µl BSA
- 7,8 µl water
- over night at 37°C
Further tasks:
- gel electrophoresis for purification
- ligation of mdnA, geneIII and mdna/geneIII (fusion gene) into pSB1C3
69th Labday 2011-08-19
Clean up of digestion products
Investigators:Katharina
Materials
- products of restriction enzyme digestion
- NucleoSpin Extract II Kit from Macherey-Nagel
miniprep of pSB1A3 and pSB1K3 clones containing either CFP or YFP and different promotors
Investigators: Nicole, Jessica, Katharina
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl
- Check concentration with NanoDrop
Results
- all concentrations were around 30 ng/µl so the preps were thrown away
- new overnight cultures have been prepared
Further Tasks:
- new miniprep
- confirmation of clones by restriction enzyme digestion
- sending for sequencing
Ligation of pSB1T3 backbones with Ara, Lac and constitutive promotors
Investigators: Nicole, Jessica, Katharina
Materials
- T4 DNA Ligase Buffer
- T4 Ligase Buffer
- different EcoRi and XbaI digested pSB1T3 backbones
- pSB1T3+YFP I clone b
- pSB1T3+YFP I clone a
- pSB1T3+YFP II plate 1 clone b
- pSB1T3+YFP II clone a
- pSB1T3+YFP I clone d: 101.8 ng/µl
- EcoRI and XbaI digested pSB1C3
- EcoRI and XbaI digested inserts
- Ara-Promotor
- constitutive promotor
- Lac-Promotor
- water
Method
- 1µl 10x T4 DNA Ligase Buffer
- 1µl T4 DNA Ligase
- 2µl backbone DNA
- 5µl insert DNA
- 1µl water
- control was prepared for the constructs by taking water instead of insert DNA
Transformation of competent RV cells with Ligation products
Investigators:Jessica, Katharina
Materials
- ligation products of:
- pSB1T3+YFPI clone b + different promotors
- pSB1T3+YFPI clone a + different promotors
- pSB1T3+YFPII plate 1 clone b + different promotors
- pSB1C3 + different promotors
- competent RV cells
Method
- transformation was done using the heatshock-protocol
- pSB1T3 clones were plated on LB+agar+Tet
- pSB1C3 clones were plated on LB+agar+Cm
- incubation over night at 37°C
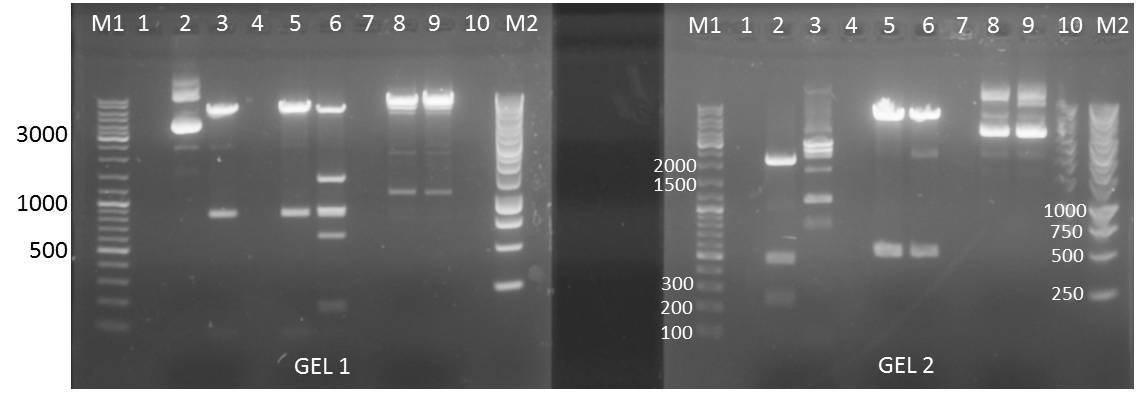
Site directed mutagenesis of 14_3C protease to remove iGEM restriction sites from the protease and introduction of iGEM restriction sites
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Aim:
- Removal of iGEM restriction sites from 14_3C protease, amplifying protease fragments with iGEM restriction sites
Materials:
- Plasmid: pGEX-3_14_3C
- Primers: (1) f_14_3C_ACCAGC, r_14_3C_iGEM_BamHI (2) r_14_3C_ACCAGC, f_14_3C_AraFusion_NgoMIV, (3) f_14_3C_tm_Xbal208_A-T, r_14_3C_tm_Xbal208_A-T (4)r_14_3C_iGEM_BamHI, f_14_3C_tm_Xbal280_A-T
Used method:
PCR
- Template: 1µl
- Nucleotides: 1µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 2µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer)
- 34,5µl of pure water
- 0,5µl TaqPol
Program:
- Denat: 3min 94°C
- 5x:
Denat: 45sec 94°C
Anneal:45sec 53°C
Extend:45sec 72°C
- 25x:
Denat: 45sec 60°C
Anneal:45sec 60°C
Extend:45sec 72°C
- Final Extend: 10min 72°C
Result:
Resolving of PCR products on preparative 2% agarose gel, exsciccion of coresponding band and gel exctraction via Nucleospin gel extract II.
Expected Fragments:
- 14_3C_mut_fragI: 153 bp
- 14_3C_mut_fragII: 66 bp
- 14_3C_mut_fragIII: 72 bp
- 14_3C_mut_fragIV: 260 bp
Further going:
- Assembly PCR of purificated products to produce NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (mutated 14_3C-Fragment)
Digestion of PCR products geneIII and mdnA for cloning into pSB1C3
Investigator: Sabine
Aim: cloning of mdnA and geneIII into pSB1C3
Time: 2011-08-19, 10:00-13:00
Material/Method:
- 30 µl PCR product (990 ng mdnA or 3200 ng geneIII)
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 0,5 µl BSA
- 4 µl NEB 10x buffer 2
- 3,5 µl water
- 1 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- cloning into pSB1C3
Digestion of PCR products geneIII and mdnA for cloning mdnA/geneIII into pSB1C3 and pARW089
Investigator: Sabine
Aim: cloning of mdnA/geneIII fusion gene into pSB1C3 and pARW089 (2 step ligation)
Time: 2011-08-19, 10:00-13:00
Material/Method:
- 30 µl / 990 ng mdnA
- 1 µl restriction enzyme AgeI
- 4 µl NEB 10x buffer 1
- 5 µl water
- 1 h, 37°C
- 30 µl / 3200 ng geneIII
- 1 µl restriction enzyme NgoMIV
- 4 µl NEB 10x buffer 4
- 5 µl water
- 1 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation of mdnA and geneIII, than ligation into pSB1C3
Digestion of PCR products geneIII and mdnA for cloning into pARW089 (3 fragment ligation)
Investigator: Sabine
Aim: cloning of mdnA and geneIII into pARW089 (3 fragment ligation)
Time: 2011-08-19, 10:00-13:00
Material/Method:
- 30 µl / 990 ng mdnA
- 2 µl restriction enzyme NarI
- 1 µl restriction enzyme AatII
- 4 µl NEB 10x buffer 4
- 3 µl water
- 1 h, 37°C
- 30 µl / 3200 ng geneIII
- 1 µl restriction enzyme NgoMIV
- 1 µl restriction enzyme AatII
- 4 µl NEB 10x buffer 4
- 4 µl water
- 1 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation into pARW089
Digestion of PCR product mdnA for cloning into pARW089
Investigator: Sabine
Aim: cloning of mdnA into pARW089
Time: 2011-08-19, 10:00-13:00
Material/Method:
- 30 µl / 990 ng mdnA
- 2 µl restriction enzyme NarI
- 1 µl restriction enzyme AatII
- 4 µl NEB 10x buffer 4
- 3 µl water
- 1 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation into pARW089, than digestion with AgeI and NgoMIV and ligation of geneIII
Analysis of sequenced pPDV100
Investigator: Sabine
Aim: control of created pPDV100
Time: 2011-08-19, 14:00-14:15
Result:
- created vector contains no mdnA
Further Tasks:
- this strategy will not be further persued
Agarose gel electrophoresis of digested mdnA and geneIII
Investigator: Sabine
Aim: purification of digested DNA fragments
Time: 2011-08-19, 15:00-17:00
Material/Method:
- digested geneIII, mdnA, pSB1C3
- 1% agarose gel, loading dye, DNA ladder mix (Fermentas)
- 100 V
Results:
- sample mdnA (NarI+AatII) has been lost (ran under the gel)
- concentration pSB1C3 (Xba+Pst): 17 ng/µl
- concentration mdnA (AgeI): 3,8 ng/µl
- concentration mdnA (XbaI+PstI): 5,5 ng/µl
- concentration geneIII (NgoMIV): 15,5 ng/µl
- concentration geneIII (XbaI+PstI): 12,2 ng/µl
- concentration geneIII (NgoMIV+AatII): 12,8 ng/µ
Further Tasks:
- repeat PCR and digestion of mdnA (NarI+AatII)
- ligations
Repeated PCR of mdnA
Investigator: Sabine
Time: 2011-08-19,17:00-19:00
Aim:
- amplification of mdnA with NarI and AgeI restriction sites
Primer:
- primer: pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII
Reaction Components:
- 2 µl Vector pARW089 (16 ng)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 39,75 µl water
Further tasks:
- purification
- digestion
Site directed mutagenesis of TEV protease to remove iGEM restriction sites from the protease and introduction of iGEM restriction sites and assembly of produced TEV-fragments
Investigators: Sascha, Paul
- Mutagenesis was performed as described before in the wiki.
- 3 reaction batches
Results:
Expected fragments (360bp and 400bp) were excissed and extracted from the gel using Nucleospin Extract Kit
- Assembly PCR of TEV fragments I and II was performed as described before in the wiki
The expected mutated complete TEV fragment was PCR purificated using nucleospin extract KIT
Amplificarion of arabinose induction system (AraC) from pBAD_iGEMexpress plasmid, produces a 1273bp fragment
Investigators: Sascha, Paul, Sebastian
Materials:
Plasmid: pBAD_iGEMexpress (Nr.4) Primers: f_AraC_HindIII_iGEM , r_AraC_NgoMIV
Used method: PCR
Template: 1µl = 7,2 ng
Nucleotides: 1 µl of 10mM ready to use dNTP mix 5µl 10x
Amplification buffer S: 5µl 25mM MgCl2 2,5µl
primers = 25pmol absolute (2,5µl of each primer = 5µl per tube) 32,5µl of pure water 0,5µl TaqPol
Program: iGEM002
Denat: 3min 94°C
5x:
Denat: 60sec 94°C
Anneal:60sec 53°C
Extend:60sec 72°C
25x:
Denat: 60sec 60°C
Anneal:60sec 60°C
Extend:60sec 72°C
Final Extend: 10min 72°C
- PCR purification using NucleoSpin Extract KIT and digestion of AraC fragment (1273bp) witth NgoMIV and HindIII
- Resolving of digested AraC fragment on 1.5% preparative agarose Gel
- The corresponding band (1273bp) was excissed and extracted from the gel using Nucleospin Extract KIT.
Expected Fragments: HindIII_iGEM_AraC_NgoMIV 1273bp
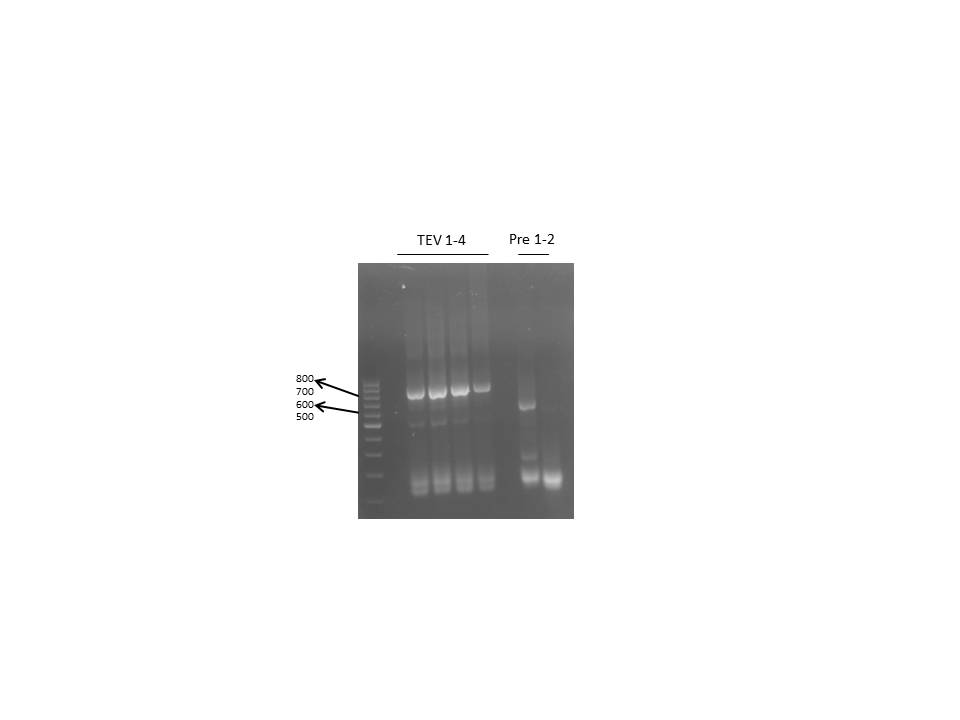
Digestion of complete mutated TEV and 14_3C fragments and 3x-ligation into TEV- or 14_3C-backbones with amplified and digested AraC fragment
Investigators: Paul, Stefan
Materials:
(1)NgoMIV_iGEM_TEV-Protease_iGEM_BamHI
(2)NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI
(3)HindIII_iGEM_AraC_NgoMIV (digested)
- digestion of proteases (1) and (2) with NgoMIV and BamHI as described before in the wiki with NgoMIV and BamHI
- resolving of digested fragments on preparative agarose gel and excission of corresponding bands. Extraction of DNA from the gel was performed using NucleoSping ExtractII KIT.
Ligation of digested proteases into digested the pJC354-14_3C/TEV vectors including the AraC fragment (3)
concentrations for ligations were calculated using gibthon ligation calculator: [http://www.gibthon.org/ligate.html CALC]
- ligations were allowed to proceed for 1h at room temperature and were immediately transformed into competent XL1-blue cells using standard transformation protocol
70th Labday 2011-08-20
miniprep of pSB1A3 and pSB1K3 clones containing either CFP or YFP and different promotors
Investigator: Jessica, Katharina
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl
- Check concentration with NanoDrop
Results
- concentrations:
- pSB1A3-CFP +Ara (clone A) 12: 342.1 ng/µl
- pSB1A3-CFP +Ara (clone B) 7: 252.5 ng/µl
- pSB1A3-CFP +Ara (clone C) 6: 356.9 ng/µl
- pSB1A3-YFP +Ara (clone A) 1: 339.2 ng/µl
- pSB1A3-YFP +Ara (clone B) 5: 379.0 ng/µl
- pSB1A3-YFP +Ara (clone C) 14: 221.4 ng/µl
- pSB1A3-CFP +Lac (clone A) 15: 319.3 ng/µl
- pSB1A3-CFP +Lac (clone B) 34: 105.5 ng/µl
- pSB1A3-CFP +Lac (clone C) 33: 135.7 ng/µl
- pSB1A3-YFP +Lac (clone A) 16: 110.6 ng/µl
- pSB1A3-YFP +Lac (clone B) 4: 340.4 ng/µl
- pSB1A3-YFP +Lac (clone C) 13: 290.6 ng/µl
- pSB1A3-CFP + const (clone A) 11: 312.2 ng/µl
- pSB1A3-CFP + const (clone B) 9: 218.0 ng/µl
- pSB1A3-CFP + const (clone C) 10: 201.7 ng/µl
- pSB1A3-YFP + const (clone C) 28: 238.0 ng/µl
- pSB1K3-CFP +Ara (clone A) 25: 175.2 ng/µl
- pSB1K3-CFP +Ara (clone B) 31: 190.2 ng/µl
- pSB1K3-CFP +Ara (clone C) 24: 193.8 ng/µl
- pSB1K3-YFP +Ara (clone A) 22: 119.6 ng/µl
- pSB1K3-YFP +Ara (clone B) 19: 149.4 ng/µl
- pSB1K3-YFP +Ara (clone C) 30: 135.1 ng/µl
- pSB1K3-CFP +Lac (clone A) 20: 138.4 ng/µl
- pSB1K3-CFP +Lac (clone B) 21: 58.3 ng/µl
- pSB1K3-CFP +Lac (clone C) 23: 122.0 ng/µl
- pSB1K3-YFP +Lac (clone A) 29: 69.3 ng/µl
- pSB1K3-YFP +Lac (clone B) 17: 153.1 ng/µl
- pSB1K3-YFP +Lac (clone C) 18: 90.9 ng/µl
- pSB1K3-CFP + const (clone A) 26: 89.8 ng/µl
- pSB1K3-CFP +const (clone B) 8: 252.5 ng/µl
- pSB1K3-CFP +const (clone C) 2: 256.6 ng/µl
- pSB1K3-YFP + const (clone A) 27: 55.7 ng/µl
- pSB1K3-YFP +const (clone B) 3: 231.9 ng/µl
- pSB1K3-YFP + const (clone C) 32: 174.9 ng/µl
- pSB1K3-YFP + const (clone C) 32: 174.9 ng/µl
- pARW089 35: 333.8 ng/µl
- pARW089 36: 326.4 ng/µl
Further Tasks:
- confirmation of clones by restriction enzyme digestion
- sending for sequencing
Gel extraction of different AraC for TEV approaches
Investigator: Stefan
Aim:
- purification of DNA
Materials:
- NucleoSpin Kit (Machery Nagel)
Method:
- DNA extraction from agarose gels protocol of NucleoSpin Kit
Results:
BILD von Sabine geschickt, von mir eingefügt
Further tasks:
Ligation with TEV and 14_3C and appropriate backbone
Ligation and transformation of 14_3C with backbone
Investigator: Paul, Stefan
Aim:
- liagte and transform 14_3C with AraC and backbone
Materials:
Transformation Protocol Using Heat Shock
1) Take chemically competent E. coli cells from –80°C freezer.
a. Use XL1-blue cells for all cloning and DNA-related tasks, BL21 cells for protein/peptide expression.
2) Turn on water bath or heat block to 42°C.
3) Competent cells should be in a 1.5 ml tube. For transforming a DNA construct, use 60 ul of competent cells. .
4) Keep tubes on ice.
5) Add DNA solution into the E.coli cell suspension, mix by flicking the tube. Incubate on ice for 15-30 min.
Note: 2 µL from a T4 DNA ligase reaction are usually sufficient
6) Put tubes into heat block at 42°C for 45 seconds.
7) Put tubes back on ice for 2 minutes to reduce damage to the E. coli cells.
8) Add 750 µL of LB or DYT (with no antibiotic!). Incubate tubes for 1 hour at 37°C and 750 rpm.
9) Spread 100 ul of the resulting culture on LB agar plates (with appropriate antibiotic added!).
Note: In case of "tricky" ligations which yield few colonies, spin down the cell suspension for 3 min at 6000 rcf (since you can't spread 800 µL), discard most of the supernatant, resuspend the bacterial pellet by pipetting and use this for spreading.
10) Grow overnight at 37 °C.
11) Pick colonies about 12-16 hours later.
Method:
Results:
check clones via colony PCR for correct insert
Further tasks:
sequencing postive clones
production of competent E. coli XL 1 blue
Investigator: Stefan
Aim:
- competent E. coli XL 1 blue
Materials:
- CaCl2
- E. coli XL1 blue
Method:
Work always sterile and cold and speedy!
- All volumes deal with the common cell line!
- The cooling-centrifuge is in the tool shed , cool down to 4°C early enough , close the lid correctly!
- Prepare early enough min. 100 Eppis (1,5µl) (per cellline) and cool down to -80°C before using
- Use Milipore-filter for sterile CaCl2 , keep cool!
- prepare 15ml LB-Medium (or DYT) with the specific antibiotic (XL1-blue? Tet, BL21 ? none!), inoculate and incubate over night
- prepare 200ml LB-Medium (or DYT) with the specific antibiotic, inoculate with 2ml of the over-night-culture. Nurture the culture until OD600 at 0,35 (0,2-0,5) (if the OD is too high, the cell won’t be competent)
- keep cell suspension in sterile falcons (50ml) 20 min on ice, then centrifuge for 20min; 4°C; 2500g
- discard supernatant, carefully resuspend on ice with 10ml cold CaCl2-solution (put a little of the 10ml solution in every falcon before!), pool every resuspended aliquot of one cell line and add 40ml CaCl2-solution (total volume 50ml). Keep 30 min on ice, then centrifuge for 20min; 4°C; 2500g
- discard supernatant, carefully resuspend pellet in 5,5ml CaCl2(80mM)/Glycerol (4:1), aliquot in Eppis á 60µl and store immediately at - 80 °C
Repeated digestion of PCR product mdnA for cloning into pARW089 (3 fragment ligation)
Investigator: Sabine
Aim: cloning of mdnA and geneIII into pARW089 (3 fragment ligation)
Time: 2011-08-20, 10:00-11:30
Material/Method:
- 30 µl / 920 ng mdnA
- 2 µl restriction enzyme NarI
- 1 µl restriction enzyme AatII
- 4 µl NEB 10x buffer 4
- 3 µl water
- 1 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation into pARW089
Agarose gel electrophoresis of digested mdnA
Investigator: Sabine
Aim: purification of digested DNA fragments
Time: 2011-08-20, 11:30-12:30
Material/Method:
- digested mdnA
- 1% agarose gel, loading dye, DNA ladder mix (Fermentas)
- 100 V
Further Tasks:
- ligations
Ligation of geneIII and mdnA into pSB1C3
Investigator: Sabine
Time: 2011-08-20, 13:00-16:00
Material/Method:
- 1 µl mdnA/XbaI+PstI (5,5 ng/µl)
- 10 µl pSB1C3/XbaI+PstI(17 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 6 µl water
- 1 h, room temperature
- 10 µl geneIII/XbaI+PstI (12,2 ng/µl)
- 1 µl pSB1C3/XbaI+PstI (17 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 6 µl water
- 1 h, room temperature
Further Tasks:
- transformation
Ligation of geneIII and mdnA to get a fusion gene
Investigator: Sabine
Time: 2011-08-20, 13:00-16:00
Material/Method:
- 10 µl mdnA/AgeI (3,8 ng/µl)
- 5,6 µl geneIII/NgoMIV (15,5 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 1,4 µl water
- 1 h, room temperature
Further Tasks:
- digestion with NarI and AatII, ligation into pARW089
- digestion with XbaI and PstI, ligation into pSB1C3
Ligation of geneIII and mdnA into pARW089 (3 fragment ligation)
Investigator: Sabine
Time: 2011-08-20, 13:00-16:00
Material/Method:
- 10 µl pARW089/NarI+AatII (60 ng/µl)
- 1 µl geneIII/NgoMIV+AatII (79 ng/µl)
- 1,5 µl mdnA/NarI+AgeI (24 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 4,5 µl water
- 1 h, room temperature
Further Tasks:
- transformation
Ligation of mdnA into pARW089
Investigator: Sabine
Time: 2011-08-20, 13:00-16:00
Material/Method:
- 10 µl pARW089/NarI+AatII (60 ng/µl)
- 1 µl mdnA/NarI+AatII (47 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 6 µl water
- 1 h, room temperature
Further Tasks:
- digestion of ligation construct with AgeI and AatII, ligation of geneIII/NgoMIV+AatII (2 step ligation)
71th Labday 2011-08-21
Transformation of competent RV cells with ligation products of pSB1T3/pSB1C3 backbones with Ara, Lac and constitutive promotors
Investigator: Katharina
Idea
- because of unclear labeling the RV cells used on 2011-08-19 could have been XL1 blue, which already have Tet resistance
Material
- pSB1T3 clones
- pSB1T3+YFPI clone b (labeled with "1")
- pSB1T3+YFPI clone a (labeled with "3")
- pSB1T3+YFPII plate I clone b (labeled with "4")
- pSB1C3
- competent RV cells
Method
- transformation was done using the heatshock-protocol
- pSB1T3 clones were plated on LB+agar+Tet
- pSB1C3 clones were plated on LB+agar+Cm
- incubation over night at 37°C
Further Tasks
- pick clones for liquid culture
- miniprep
- restriction enzyme digestion for confirmation
- send for sequencing
miniprep of pSB1T3 clones containing CFP and YFP, respectively
Investigators: Katharina
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl
- Check concentration with NanoDrop
Results:
- pSB1T3+YFP I clone b: 690.6 ng/µl
- pSB1T3+YFP I clone c: 576.3 ng/µl
- pSB1T3+YFP I clone a: 646.9 ng/µl
- pSB1T3+YFP II plate 1 clone b: 599.8 ng/µl
- pSB1T3+YFP II clone a: 562.8 ng/µl
- pSB1T3+YFP II plate 2 clone d: 569.1 ng/µl
- pSB1T3+YFP II plate 2 clone c: 415.0 ng/µl
- pSB1T3+YFP I clone d: 559.9 ng/µl
Digest of pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL
Investigator: Sascha, Sebastian
Aim:
- Digestion of vector pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL for ligation with AraC and 14_3C protease
Materials:
- 4 µl of 3 different fractions of pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL (approx. 1,2-1,5 µg DNA)
- Restriction enzymes BamHI HF and HindIII (purchased form NEB)
- Buffer 4 (purchased from NEB)
Method:
- Standard digestion protocol for plasmid DNA
Results:
- 3 different digested vector fractions of pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL for ligation with AraC and 14_3C protease
Further tasks:
- purification via gel purification (Kit from Macherey-Nagel)
Output:
- pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL vector fraction 1, c= ng/ml
- pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL vector fraction 2, c= ng/ml
- pJC354_ssTorA_NheI_CS-14_3C_XhoI_blaFL vector fraction 3, c= ng/ml
Assembly PCR of different TEV mutagenesis fraction
Investigator: Sascha, Sebastian
Aim:
- Assembly of the different fractions of mutated TEV-fragments
Materials:
- TEV mutagenesis fragment I (fraction I, II, III, IV)
- TEV mutagenesis fragment II (fraction I, II, III, IV)
- Primer 1: f_TEV_AraFusion (43)
- Primer 2: r_TEV_iGEM_BamHI (46)
- Taq-Polymerase (purchased from Genaxxon)
- 10x polymerase buffer (purchased from Genaxxon)
- 10 mM (each) dNTPs (purchased from Genaxxon)
- double destilled water
- 25 mM MgCl2
Method:
PCR
- Template 1: 1µl TEV mutagenesis fragment I (fraction I-IV)
- Template 2: 1µl TEV mutagenesis fragment II (fraction I-IV)
- Nucleotides: 1µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 2µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer)
- 33,5µl of pure water
- 0,5µl TaqPol
Program:
- Denat: 3min 94°C
- 5x:
Denat: 45sec 94°C
Anneal:45sec 53°C
Extend:45sec 72°C
- 25x:
Denat: 45sec 60°C
Anneal:45sec 60°C
Extend:45sec 72°C
- Final Extend: 10min 72°C
Template batches:
- TEV mutagenesis fragment I fraction I and TEV mutagenesis fragment II fraction I - fraction 1
- TEV mutagenesis fragment I fraction I and TEV mutagenesis fragment II fraction II - fraction 2
- TEV mutagenesis fragment I fraction I and TEV mutagenesis fragment II fraction III - fraction 3
- TEV mutagenesis fragment I fraction I and TEV mutagenesis fragment II fraction IV - fraction 4
- TEV mutagenesis fragment I fraction II and TEV mutagenesis fragment II fraction I - fraction 5
- TEV mutagenesis fragment I fraction II and TEV mutagenesis fragment II fraction II - fraction 6
- TEV mutagenesis fragment I fraction II and TEV mutagenesis fragment II fraction III - fraction 7
- TEV mutagenesis fragment I fraction II and TEV mutagenesis fragment II fraction IV - fraction 8
- TEV mutagenesis fragment I fraction III and TEV mutagenesis fragment II fraction I - fraction 13
- TEV mutagenesis fragment I fraction III and TEV mutagenesis fragment II fraction II - fraction 14
- TEV mutagenesis fragment I fraction III and TEV mutagenesis fragment II fraction III - fraction 15
- TEV mutagenesis fragment I fraction IV and TEV mutagenesis fragment II fraction IV - fraction 16
- TEV mutagenesis fragment I fraction IV and TEV mutagenesis fragment II fraction I - fraction 9
- TEV mutagenesis fragment I fraction IV and TEV mutagenesis fragment II fraction II - fraction 10
- TEV mutagenesis fragment I fraction IV and TEV mutagenesis fragment II fraction III - fraction 11
- TEV mutagenesis fragment I fraction IV and TEV mutagenesis fragment II fraction IV - fraction 12
Results:
- 16 different assembled TEV protease fractions with side directed mutagenesis to remove iGEM RS in nucleotide sequence
Further tasks:
- control of fragments with analytical agarose gelelctrophoresis
- PCR purification of fractions with the complete assembled TEV-Protease
- digest of TEV protease fractions with restriction enzymes NgoMIV and PstI and ligation with AraC into the digested (with EcoRI and PstI) vector pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
- transformation of competent E.coli XL1 blue cells with ligated fragments, picking clones, colony PCR, mini-prep of plasmid, sequencing plasmid and survival test with positiv clones
Output:
- 5 assembled TEV proteases
- TEV batch 2 c=
- TEV batch 2 c=
- TEV batch 3 c=
- TEV batch 3 c=
- TEV batch 7 c=
- TEV batch 7 c=
- TEV batch 10 c=
- TEV batch 10 c=
- TEV batch 16 c=
- TEV batch 16 c=
72th Labday 2011-08-22
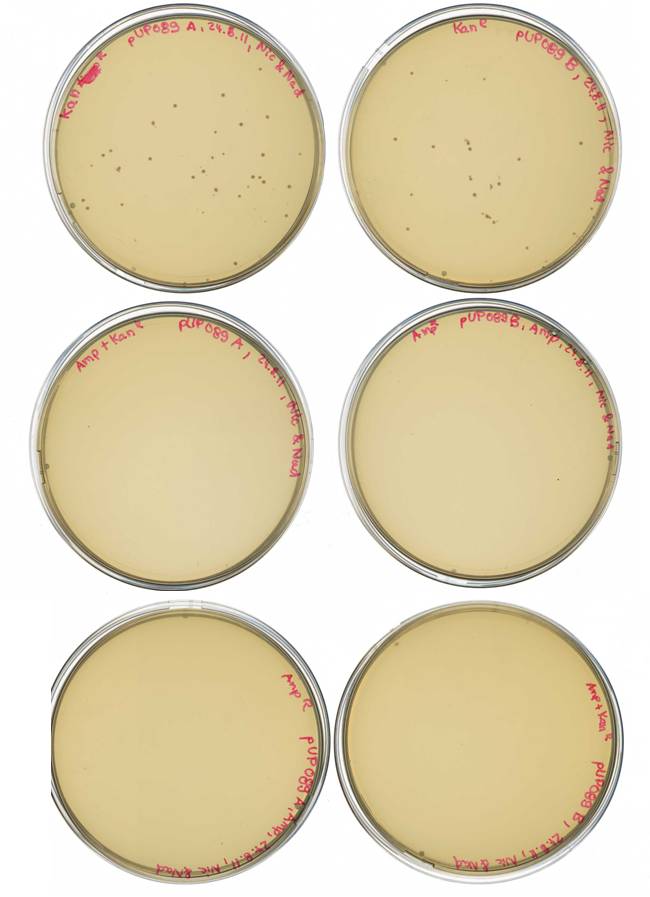
Elimination of ampicillin resistance in pARW089
Investigators: Nadine, Jessica, Steffi, Nicole
Time: 2011-08-22
Aim:
- generation of pUP089 from pARW089, vector without ampicillin resistance
- for mdnA library and later screening
idea:
- digest w/ AvaII and religation
New primer ordered for cloning of geneIII into pARW089 (without mdnA)
Investigators: Sabine
Aim:
- generation of pARW089 containing only geneIII but not mdnA gene
- mdnA library could cloned into this vector for screening
Time: 16:00-16:35
Material/Method:
- Geneious
Result:
- new forward primer for geneIII containing NarI and NgoMIV restriction site and myc
- PCR product geneIII can cloned with NarI and AatII into pARWo89
Further tasks:
- PCR, digestion and ligation
Digestion of mdnA/geneIII fusion gene for cloning into pARW089 (2 step ligation)
Investigator: Sabine
Aim: cloning of mdnA/geneIII fusion gene into pARW089
Time: 2011-08-22, 16:30-17:30
Material/Method:
- 30 µl / 500 ng mdnA/geneIII
- 2 µl restriction enzyme NarI
- 1 µl restriction enzyme AatII
- 4 µl NEB 10x buffer 4
- 3 µl water
- 1 h, 37°C
Further Tasks:
- ligation into pARW089
Digestion of mdnA/geneIII fusion gene for cloning into pSB1C3 (2 step ligation)
Investigator: Sabine
Aim: cloning of mdnA/geneIII fusion gene into pSB1C3
Time: 2011-08-22, 16:30-17:30
Material/Method:
- 30 µl / 500 ng geneIII
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 4 µl NEB 10x buffer 2
- 4 µl water
- 0,4 µl BSA
- 1 h, 37°C
Further Tasks:
- ligation into pSB1C3
Ligation of mdnA/geneIII into pARW089
Investigator: Sabine
Time: 2011-08-22, 18:00-19:00
Material/Method:
- 10 µl pARW089/NarI+AatII (60 ng/µl)
- 8 µl mdnA/geneIII (16 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 1 h, room temperature
Further Tasks:
- transformation
Ligation of mdnA/geneIII into pSB1C3
Investigator: Sabine
Time: 2011-08-22, 18:00-19:00
Material/Method:
- 8 µl pARW089/NarI+AatII (60 ng/µl)
- 9 µl mdnA/geneIII (16 ng/µl)
- 2 µl 10x T4 ligase buffer
- 1 µl T4 ligase
- 1 h, room temperature
Further Tasks:
- transformation
Transformation of generated vector constructs in E. coli
Investigators: Sabine
Aim: amplification and control of generated vectors
- mdnA, geneIII or mdnA/geneIII in pSB1C3
- mdnA or mdnA/geneIII in pARW089
Time: 20:30-22:00
Method:
- addition of 4 µl ligation reaction to XL1-blue cells
- incubation 15 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml kanamycin (pARW089) or chloramphenicol (pPSB1C3)
- storage over night at 37°C
Further tasks:
control cell clones
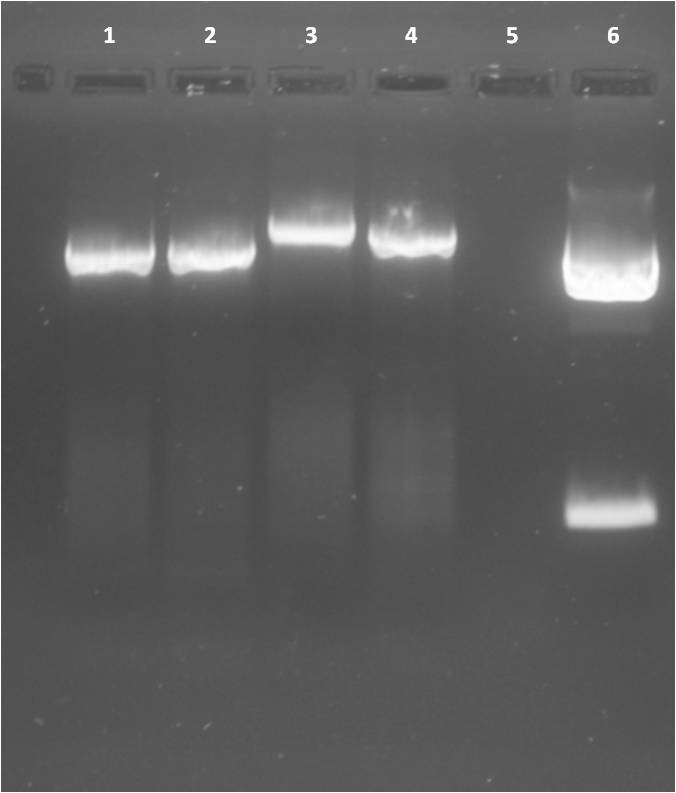
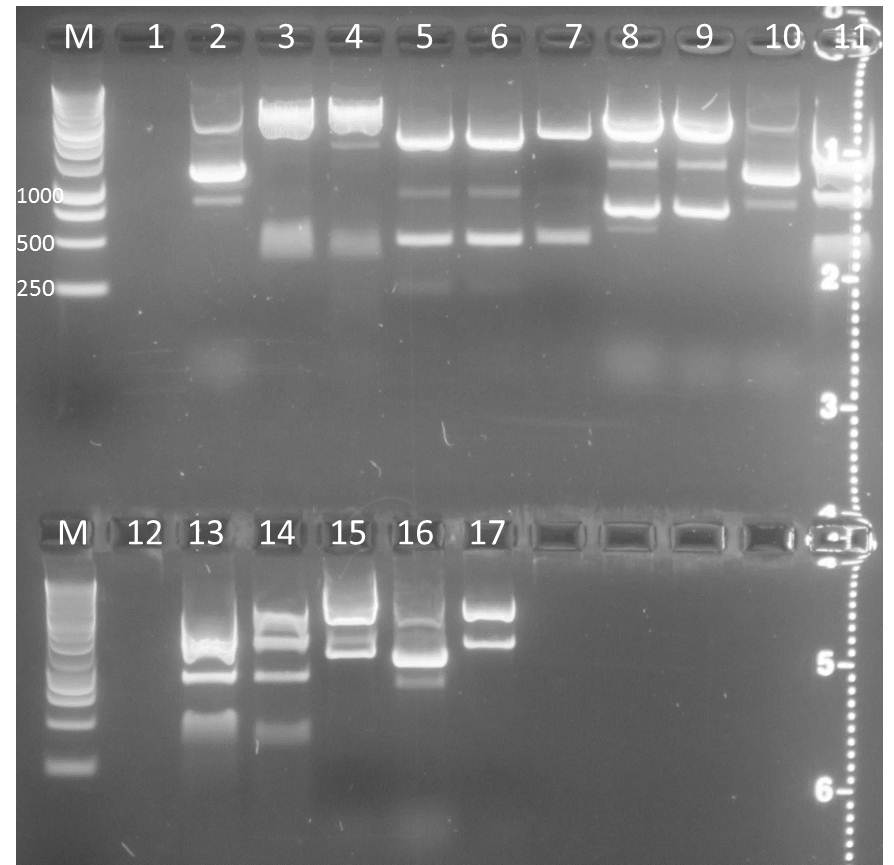
Digest of pSB1C3, pSB1T3 clones containing CFP and YFP, respectively as well as PCR products of promoters
Investigators: Jessica
Time: 2011-08-22
Materials
- pSB1C3 (BBa_K40304_Affibody_MiddleLinker) from
- pSB1T3+YFP I clone b and pSB1T3+YFP II clone a from 2011-08-21 (Katharina)
- PCR fragments: Arabinose, IPTG-inducible, constitutive
Method:
- Reaction mix:
| pSB1C3 | pSB1T3+YFP I clone b | pSB1T3+YFP II clone a | PCR fragments | |
|---|---|---|---|---|
| DNA | 6 | 3 | 3 | 20 |
| 10x Buffer 4 NEB | 2 | 2 | 2 | 3 |
| XbaI | 1 | 1 | 1 | 1 |
| EcoRI | 1 | 1 | 1 | 1 |
| 100x BSA | 0.2 | 0.2 | 0.2 | 0.3 |
| H2O | 9.8 | 12.8 | 12.8 | 4.8 |
| total | 20 | 20 | 20 | 30 |
- at 37°C for approx. 3 h
- heat inactivation at 65°C for 20 min
Agarose gel
Gel extraction:
- using Promega- Wizard SV Gel and PCR Clean Up System
- elute in 30 µl nuclease-free water
Dephosphorylation of pSB1C3:
- using Promega TSAP
- adding 3 µl Buffer and 1 µl TSAP
- incubate at 37°C for 15 min
- heat inactivaton at 74°C for 15 min
Results:
73th Labday 2011-08-23
overnight culture of picked E. coli clones transformed with pPDV
Investigators: Sabine
Aim: amplification and purification of generated phage display vector pPDV089 for test digestion and sequencing
Time: 11:00-13.00
Method/Materials:
- 10 clones pARW089+mdnA/geneIII from 3 ragment ligation
- 10 clones pARW089+mdnA/geneIII from 2 step ligation
- 5 clones pARW089+mdnA
- 5 clones pSB1C3+mdnA
- 5 clones pSB1C3+geneIII
- 5 clones pSB1C3+mdnAIgeneIII
- 5 ml LB medium per clone containining 25 µg/ml chloramphenicol (pSB1C3) or kanamycin (pARW089)
- storage over night at 37°C and 800 rpm
Further tasks:
plasmid preparation, test digestion and sequencing
PCR: BioBrick mdnABC, mdnC, mdnE, mdnDE, mdnBC
Time: 2011-08-23, 8:00-11:30
Investigators: Nadine, Nicole
Materials
- vector: pARW089, 20.8.11, 333.4 ng/µl
- Phusion HF Polymerase, NEB
- Phusion HF Buffer
- dNTPs
- water
- Primer:
| # | Primer |
|---|---|
| 57 | pf_mdnABC_EcoRI_NotI_XbaI 12.08. |
| 58 | pf_mdnB_EcoRI_NotI_XbaI 12.08. |
| 59 | pf_mdnC_EcoRI_NotI_XbaI 12.08. |
| 60 | pf_mdnD_EcoRI_NotI_XbaI 12.08. |
| 61 | pf_mdnE_EcoRI_NotI_XbaI 12.08. |
| 62 | pr_mdnABC_SpeI_NotI_PstI 12.08. |
| 63 | pr_mdnB_SpeI_NotI_PstI 12.08. |
| 64 | pr_mdnD_SpeI_NotI_PstI 12.08. |
| 65 | pr_mdnE_SpeI_NotI_PstI 12.08. |
Protocol:
- Reaction mix
- 2 µl pARW089 (diluted 1:100)
- 1 µl dNTPs (10 mM)
- 2.5 µl Primer forward (10 mM)
- 2.5 µl Primer backward (10 mM)
- 10 µl Buffer (5x)
- 0.5 µl Phusion HF Ploymerase (2U/µl)
- 31.5 µl water
- total volume: 50 µl
- Master mix
- 12 µl pARW089 (diluted 1:100)
- 6 µl dNTPs (10 mM)
- 60 µl Buffer (5x)
- 3 µl Phusion HF Ploymerase (2U/µl)
- 189 µl water
- Primer
- mdnABC: 57, 62
- mdnC: 59, 62
- mdnE: 61, 65
- mdnDE: 61, 64
- mdnBC: 58, 62
2. PCR programs
- IGBIO02 for mdnABC, mdnC
- first steps: 10x
- second steps: 20x
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 98°C | 10 s |
| Annealing | 59°C | 30 s |
| Extension | 72°C | 20 s |
| DenaturationII | 98°C | 10 s |
| AnnealingII | 72°C | 30 s |
| ExtensionII | 72°C | 20 s |
| Final extension | 72°C | 10 min |
- IGBIO03 for mdnE, mdnDE, mdnBC
- first steps: 10x
- second steps: 20x
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 98°C | 10 s |
| Annealing | 52°C | 30 s |
| Extension | 72°C | 60 s |
| DenaturationII | 98°C | 10 s |
| AnnealingII | 72°C | 30 s |
| ExtensionII | 72°C | 60 s |
| Final extension | 72°C | 10 min |
Results:
- Output:
- mdnABC, Nad & Nic, 23.8.11
- mdnC, Nad & Nic, 23.8.11
- mdnE, Nad & Nic, 23.8.11
- mdnDE, Nad & Nic, 23.8.11
- mdnBC, Nad & Nic, 23.8.11
Digest of pSB1T3+YFPI/II vectors for ligation w/ promotors
Time: 2011-08-23, 9:00-
Investigators: Nadine, Nicole, Katharina
Materials
- vectors from 2011-08-21 (Katharina):
- pSB1T3+YFP I clone b: 690.6 ng/µl
- pSB1T3+YFP I clone c: 576.3 ng/µl
- pSB1T3+YFP I clone a: 646.9 ng/µl
- pSB1T3+YFP II plate 1 clone b: 599.8 ng/µl
- pSB1T3+YFP II clone a: 562.8 ng/µl
- pSB1T3+YFP II plate 2 clone d: 569.1 ng/µl
- pSB1T3+YFP II plate 2 clone c: 415.0 ng/µl
- pSB1T3+YFP I clone d: 559.9 ng/µl
- EcoRI HF
- XbaI
- BSA (10x)
- Buffer 4
- water
Protocol:
- Reaction mix
- 2 µl DNA
- 1 µl EcoRI
- 1 µl XbaI
- 0.3 µl BSA
- 3 µl Buffer 4 (10x)
- 22.7 µl water
- total volume: 30 µl
- incubation at 37°C for 2 hrs
Agarose gel
Investigator: Jessica
- Gel extraction using NucleoSpin ExtractII (Macherey-Nagel)
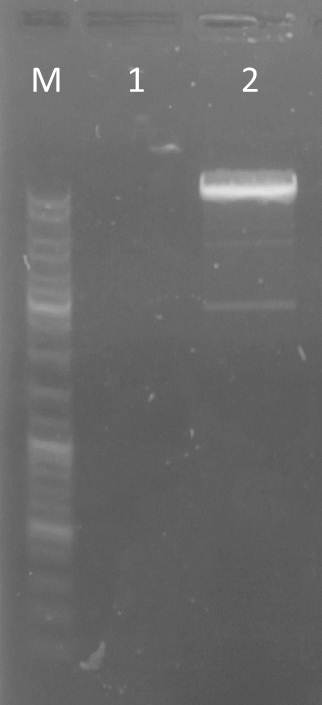
- no excision of pSB1T3+YFP II plate 2 clone c because of no visible band
Results:
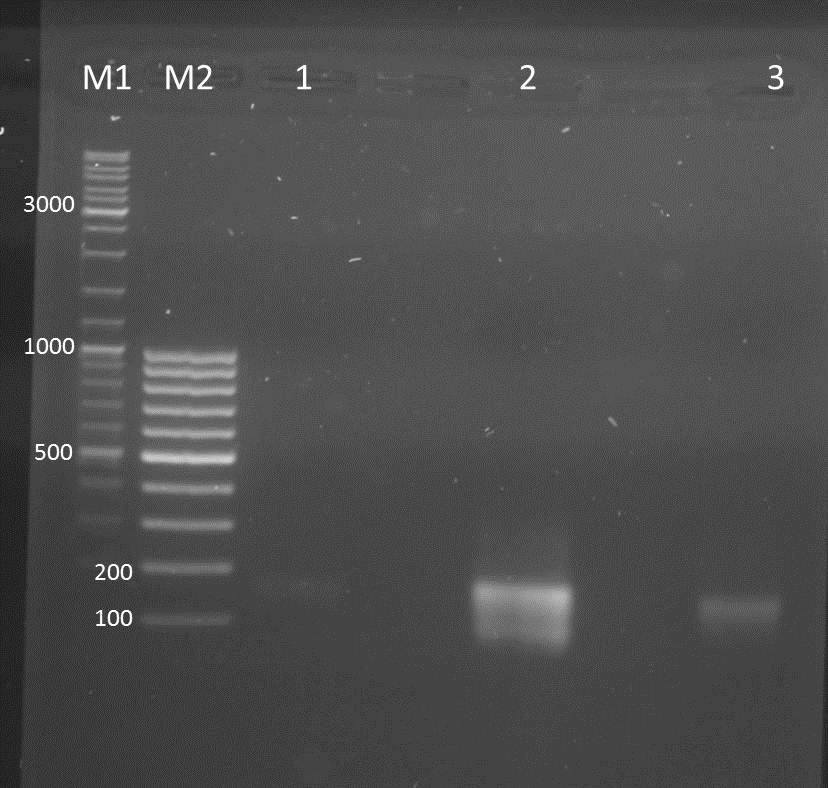
Digest of amplified 14_3C protease (assembled fragments after sidedirected mutagenesis), amplified AraC (from pBAD_iGEM_express mVenus), plasmid pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL and plasmid pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL
Investigator: Sebastian, Sascha
Material:
- Restriction enzymes NgoMIV, HindIII, BamHI, PstI-HF, EcoRI-HF (purchased from NEB)
- digest buffer 2 and 4 (puchased from NEB)
- BSA (purchased from NEB)
- plasmid pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL
- plasmid pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL
- PCR amplified AraC-fragment
- PCR amplified and mutated 14_3C protease
Methode:
Standard protocoll for DNA digest.
Reaction batches:
- Batch 1: 21.7 µl 14_3C fraction I (c=157.0 ng/µl), 2.5 µl NgoMIV, 2.5 µl BamHI, 0.3 µl BSA, 3 µl Buffer 4
- Batch 2: 21.7 µl 14_3C fraction II(c=157.5 ng/µl), 2.5 µl NgoMIV, 2.5 µl BamHI, 0.3 µl BSA, 3 µl Buffer 4
- Batch 3: 22.0 µl AraC (c=175.0 ng/µl), 2.5 µl NgoMIV, 2.5 µl HindIII, 3 µl Buffer 2
- Batch 4: 22.0 µl AraC (c=175.0 ng/µl), 2.5 µl NgoMIV, 2.5 µl EcoRI-HF, 3 µl Buffer 4
- Batch 5: 6,9 µl plasmid pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL II (c= ng/µl), 1.0 µl HindIII, 2.5 µl BamHI, 0.1 µl BSA, 1 µl Buffer 2
- Batch 6: 6,9 µl plasmid pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL VII (c= ng/µl), 1.0 µl HindIII, 2.5 µl BamHI, 0.1 µl BSA, 1 µl Buffer 2
- Batch 7: 7.0 µl plasmid pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL (c= ng/µl), 1.0 µl EcoRI-HF, 1.0 µl PstI-HF, 1.0 µl Buffer 4
Further tasks:
All plasmid fractions should be purified with 1.0% agarose gel, all other fragments with 1.5% agarose gel and extracted with Macherey-Nagel Nucleo SpinII Kit.
- Ligation of fragments in different combinations and transformation of competent E.coli XL1 blue cells.
Output:
- digested fragments of AraC, 14_3C protease and plasmid backbones for transformation (AraC + 14_3C + "14_3C-backbone", AraC + TEV + "TEV-backbone")
1) EcoRI_AraC_NgoMIV
2) HindIII_AraC_NgoMIV
3) NgoMIV_14_3C_BamHI Batch 1
4) NgoMIV_14_3C_BamHI Batch 2
5) BamHI_pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL_HindIII Batch 1
6) BamHI_pJC354_ssTorA_XhoI_CS-143C_NheI_blaFL_HindIII Batch 2
7) BamHI_pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL_HindIII
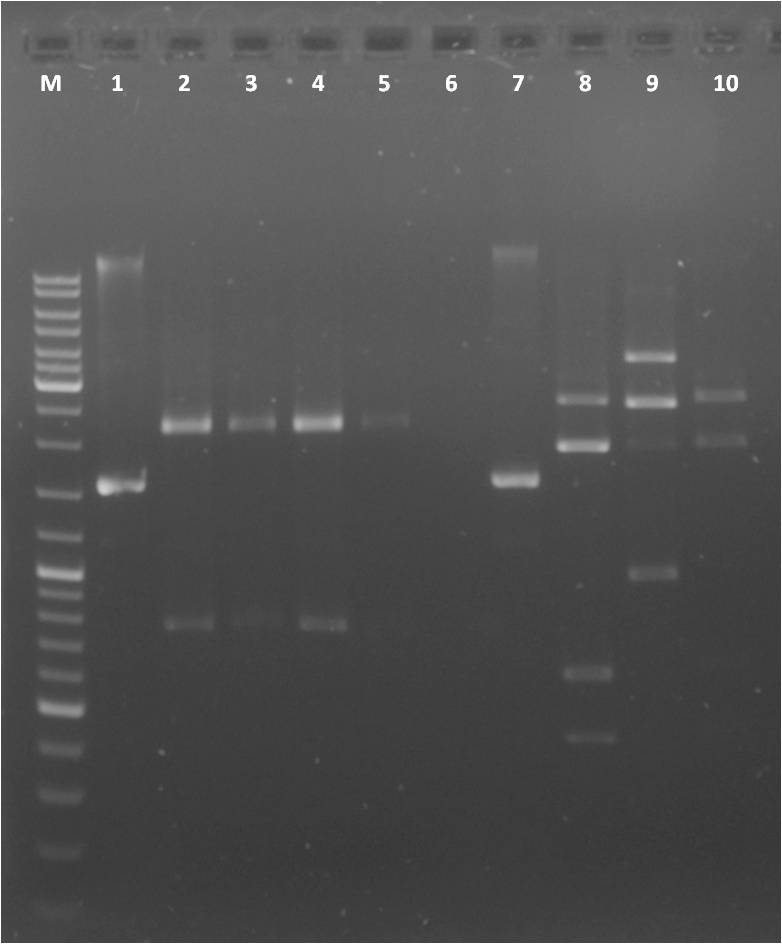
Agarose Gel of PCR products for BioBricks mdnABC, mdnBC, mdnC, mdnC, mdnD, mdnDE, mdnE and PCR purification
Investigators: Nadine, Nicole, Katharina, Jessica
Time: 2011-08-23, 13:00-16:00
Materials:
- PCR products
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Production of one 1 % agarose gel
- 2 x 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
- Add µl Loading dye to each 2 µl sample
- 12 µl DNA Ladder Mix
- Running conditions: 110 V, approx. 45 min
Loading of gel
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker | 12 | |
| 1 | mdnABC | - | 2492 |
| 2 | mdnB | - | 1021 |
| 3 | mdnBC | - | 2045 |
| 4 | mdnC | - | 1007 |
| 5 | mdnD | - | 600 |
| 6 | mdnDE | - | 2800 |
| 7 | mdnE | - | 2100 |
Result gel 1:
- all PCR products appear as expected, except mdnB
- purification of PCR products w/ Machery and Nagel PCR purification kit
- Output:
- PCR pur., mdnABC, 48.1 ng/µl, Kat, 23.8.11
- PCR pur., mdnBC, 70.1 ng/µl, Kat, 23.8.11
- PCR pur., mdnC, 62 ng/µl, Kat, 23.8.11
- PCR pur., mdnD, 67.1 ng/µl, Kat, 23.8.11
- PCR pur., mdnDE, 61.8 ng/µl, Kat, 23.8.11
- PCR pur., mdnE, 68.7 ng/µl, Kat, 23.8.11
Further task:
- repeat PCR for mdnB
- digestion of other PCR products
- ligation w/ pSB1C3
Test digest of pSB1A3+CFP/YFP and pSB1K3+CFP/YFP vectors w/ promotors
Investigators: Steffi, Nadine, Nicole
Materials
- vectors from 2011-08-20 (Katharina):
- pSB1A3-CFP +Ara (clone A) 12: 342.1 ng/µl
- pSB1A3-CFP +Ara (clone B) 7: 252.5 ng/µl
- pSB1A3-CFP +Ara (clone C) 6: 356.9 ng/µl
- pSB1A3-YFP +Ara (clone A) 1: 339.2 ng/µl
- pSB1A3-YFP +Ara (clone B) 5: 379.0 ng/µl
- pSB1A3-YFP +Ara (clone C) 14: 221.4 ng/µl
- pSB1A3-CFP +Lac (clone A) 15: 319.3 ng/µl
- pSB1A3-CFP +Lac (clone B) 34: 105.5 ng/µl
- pSB1A3-CFP +Lac (clone C) 33: 135.7 ng/µl
- pSB1A3-YFP +Lac (clone A) 16: 110.6 ng/µl
- pSB1A3-YFP +Lac (clone B) 4: 340.4 ng/µl
- pSB1A3-YFP +Lac (clone C) 13: 290.6 ng/µl
- pSB1A3-CFP + const (clone A) 11: 312.2 ng/µl
- pSB1A3-CFP + const (clone B) 9: 218.0 ng/µl
- pSB1A3-CFP + const (clone C) 10: 201.7 ng/µl
- pSB1A3-YFP + const (clone C) 28: 238.0 ng/µl
- pSB1K3-CFP +Ara (clone A) 25: 175.2 ng/µl
- pSB1K3-CFP +Ara (clone B) 31: 190.2 ng/µl
- pSB1K3-CFP +Ara (clone C) 24: 193.8 ng/µl
- pSB1K3-YFP +Ara (clone A) 22: 119.6 ng/µl
- pSB1K3-YFP +Ara (clone B) 19: 149.4 ng/µl
- pSB1K3-YFP +Ara (clone C) 30: 135.1 ng/µl
- pSB1K3-CFP +Lac (clone A) 20: 138.4 ng/µl
- pSB1K3-CFP +Lac (clone B) 21: 58.3 ng/µl
- pSB1K3-CFP +Lac (clone C) 23: 122.0 ng/µl
- pSB1K3-YFP +Lac (clone A) 29: 69.3 ng/µl
- pSB1K3-YFP +Lac (clone B) 17: 153.1 ng/µl
- pSB1K3-YFP +Lac (clone C) 18: 90.9 ng/µl
- pSB1K3-CFP + const (clone A) 26: 89.8 ng/µl
- pSB1K3-CFP +const (clone B) 8: 252.5 ng/µl
- pSB1K3-CFP +const (clone C) 2: 256.6 ng/µl
- pSB1K3-YFP + const (clone A) 27: 55.7 ng/µl
- pSB1K3-YFP +const (clone B) 3: 231.9 ng/µl
- pSB1K3-YFP + const (clone C) 32: 174.9 ng/µl
- pSB1K3-YFP + const (clone C) 32: 174.9 ng/µl
- BamHI
- AatII
- HpaI
- HincII
- NotI
- BSA (10x)
- Buffer 3
- Buffer 4
- water
Protocol:
- Reaction mix (Arabinose)
- 0.5 µl DNA
- 1 µl BamHI
- 1 µl AatII
- 0.3 µl BSA
- 3 µl Buffer 4 (10x)
- 24.2 µl water
- Reaction mix (Lac)
- 0.5 µl DNA
- 1 µl HpaI
- 1 µl AatII
- 3 µl Buffer 4 (10x)
- 24.5 µl water
- Reaction mix (constitutive)
- 0.5 µl DNA
- 1 µl HincII
- 1 µl NotI
- 0.3 µl BSA
- 3 µl Buffer 3 (10x)
- 24.2 µl water
- total volume each: 30 µl
- incubation at 37°C for 1 hrs
further tasks:
- gel electrophoresis
- ligation
- transformation
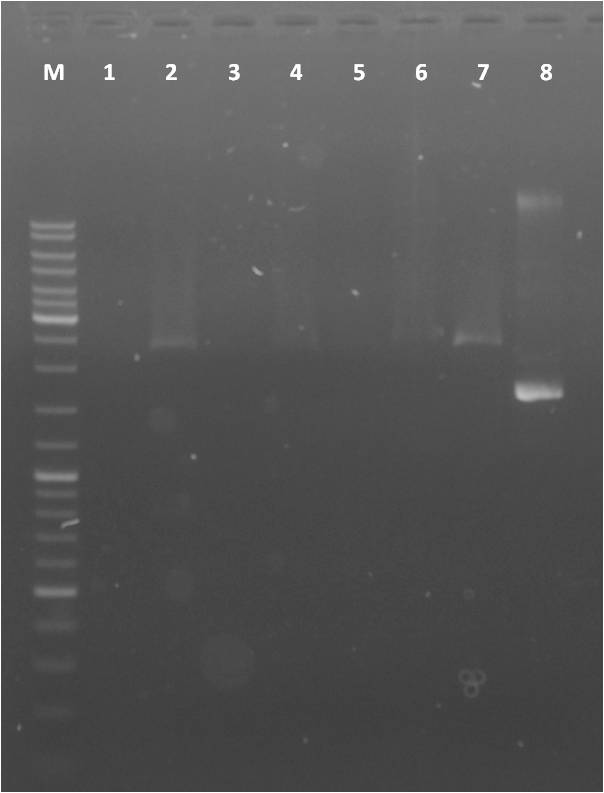
Agarose Gel of pSB1A3+CFP/YFP and pSB1K3+CFP/YFP
Investigators: Steffi, Katharina, Nadine
Materials:
- digestion products
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Production of one 1 % agarose gel
- 1 x 1 % gel: 1,0 g agarose in 100 ml 1x TAE buffer
- Adding 4 µl gel red to gel
Loading gels and running
- Add 5 µl Loading dye to each 2 µl sample
- 7 µl DNA Ladder Mix
- Running conditions: 110 V, approx. 45 min
Loading of gel
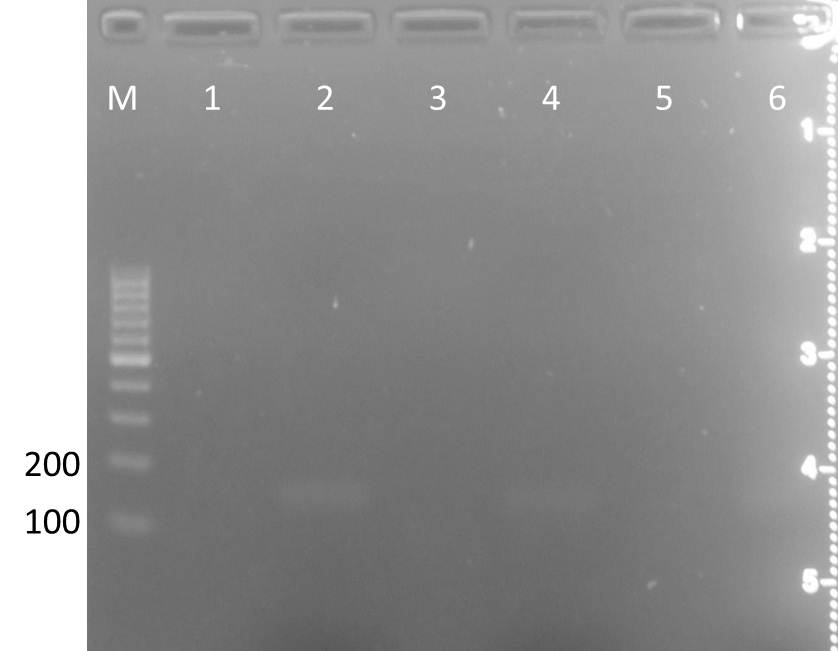
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker | 7 | |
| 1 | pSB1A3-CFP +Ara (clone A) | 15 | |
| 2 | pSB1A3-CFP +Ara (clone B) | 15 | |
| 3 | pSB1A3-CFP +Ara (clone C) | 15 | |
| 4 | pSB1A3-YFP +Ara (clone A) | 15 | |
| 5 | pSB1A3-YFP +Ara (clone B) | 15 | |
| 6 | pSB1A3-YFP +Ara (clone C) | 15 | |
| 7 | pSB1K3-CFP +Ara (clone A) | 15 | |
| 8 | pSB1K3-CFP +Ara (clone B) | 15 | |
| 9 | pSB1K3-CFP +Ara (clone C) | 15 | |
| 10 | pSB1K3-YFP +Ara (clone A) | 15 | |
| 11 | pSB1K3-YFP +Ara (clone B) | 15 | |
| 12 | pSB1K3-YFP +Ara (clone C) | 15 | |
| M | marker | 7 | |
| 13 | pSB1A3-CFP + const (clone A) | 15 | |
| 14 | pSB1A3-CFP + const (clone B) | 15 | |
| 15 | pSB1A3-CFP + const (clone C) | 15 | |
| 16 | pSB1A3-YFP + const (clone C) | 15 | |
| 17 | pSB1K3-CFP + const (clone A) | 15 | |
| 18 | pSB1K3-CFP + const (clone B) | 15 | |
| 19 | pSB1K3-CFP + const (clone C) | 15 | |
| 20 | pSB1K3-YFP + const (clone A) | 15 | |
| 21 | pSB1K3-YFP + const (clone B) | 15 | |
| 22 | pSB1K3-YFP + const (clone C) | 15 | |
| M | marker | 7 | |
| 23 | pSB1A3-CFP +Lac (clone A) | 15 | |
| 24 | pSB1A3-CFP +Lac (clone B) | 15 | |
| 25 | pSB1A3-CFP +Lac (clone C) | 15 | |
| 26 | pSB1A3-YFP +Lac (clone A) | 15 | |
| 27 | pSB1A3-YFP +Lac (clone B) | 15 | |
| 28 | pSB1A3-YFP +Lac (clone C) | 15 | |
| 29 | pSB1K3-CFP +Lac (clone A) | 15 | |
| 30 | pSB1K3-CFP +Lac (clone B) | 15 | |
| 31 | pSB1K3-CFP +Lac (clone C) | 15 | |
| 32 | pSB1K3-YFP +Lac (clone A) | 15 | |
| 33 | pSB1K3-YFP +Lac (clone B) | 15 | |
| 34 | pSB1K3-YFP +Lac (clone C) | 15 |
Result gel:
- sample ... used for sequencing
further tasks:
- sequencing
Agarose Gel of pARW089 Ava II A (3µl DNA)/ B (4µl DNA) (= pUP089)
Investigators: Niels, Nadja
Materials:
- digestion products
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Production of one 0,8 % agarose gel
- 1 x 0,8 % gel: 0,4 g agarose in 50ml 1x TAE buffer
- Adding 2 µl gel red to gel
Loading gels and running
- Add 6 µl Loading dye to each 30 µl sample
- 20 µl DNA Ladder Mix 1:10 diluted
- Running conditions: 100 V, approx. 50 min
Loading of gel
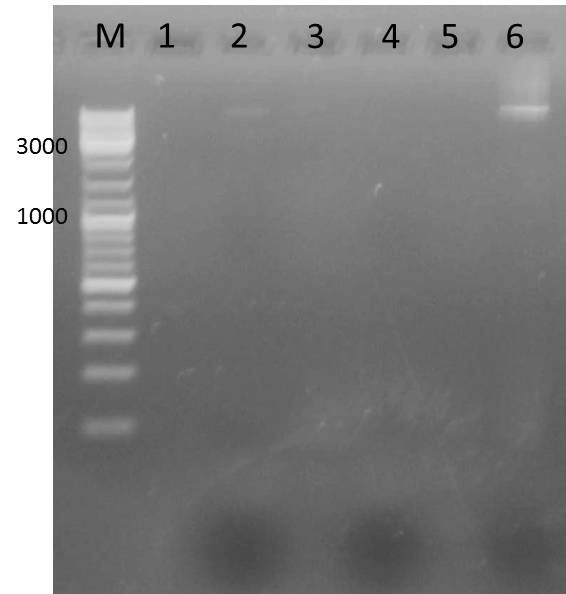
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker | 20 | |
| 1 | pARW089 Ava II A | 36 | over 10kbp |
| 2 | pARW089 Ava II B (4µl DNA) | 36 | over 10kbp |
Result gel:
- sample used for gel extraction (Promega Kit Wizard SV Gel and PCR Clean Up System)
- sample used for ligation
Further tasks:
- transformation
Ligation of pSB1C3 and pSB1T3-YFPI/II with promoters and ligation of pARW089 AvaII
Investigators: Jessica
Materials:
- T4 Ligase (Promega)
- 10x T4 Ligase Buffer (Promega)
- digested, gel purified, dephoshorylated vectors: pSB1C3, pSB1T3-YFP I clone c, pSB1T3-YFP II plate 2 clone d
- Inserts: Ara promoter, Lac promoter, constitutive promoter
Method:
- 10 µl reaction for pSB vectors:
- 1 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 6 µl vector
- 2 µl insert
- 10 µl reaction for pARW089 vector:
- 1 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 8 µl vector
- at 14°C overnight
Results:
- 14 ligation products
- pSB1C3 + Ara
- pSB1C3 + Lac
- pSB1C3 + const
- pSB1C3 + H2O
- pSB1T3-YFP II + Ara
- pSB1T3-YFP II + Lac
- pSB1T3-YFP II + const
- pSB1T3-YFP II + H2O
- pSB1T3-YFP I + Ara
- pSB1T3-YFP I + Lac
- pSB1T3-YFP I + const
- pSB1T3-YFP I + H2O
- pSB1T3-YFP I + H2O
- pARW089 AvaII A = pUP089 A
- pARW089 AvaII B = pUP089 B
Further tasks:
- transformation
Over night culture of several pSB1A3+CFP/YFP and pSB1K3+CFP/YFP with promotor
Time: 2011-08-23, 18:00-22:30
Investigators: Niels, Jessica, Nadja
Materials
- 5 ml LB media
- 5 µl antibotic (KAN/AMP - Stock)
- clone
- KAN YFP ava clone IV (from plate)
- KAN YFP ava clone V (from plate)
- KAN YFP ava clone VI (from plate)
- KAN CFP const clone I (from pre-culture)
- KAN CFP const clone II (from pre-culture)
- KAN CFP const clone III (from pre-culture)
- KAN YFP const clone I (from pre-culture)
- KAN YFP const clone II (from pre-culture)
- KAN YFP const clone III (from pre-culture)
- AMP CFP const clone I (from pre-culture)
- AMP CFP const clone II (from pre-culture)
- AMP CFP const clone III (from pre-culture)
- AMP YFP const clone I (from pre-culture)
conditions
- 37 °C
- 250 rpm
- over night
further tasks:
- miniprep
- digest
- analytic gel
sequencing of
Time: 2011-08-23, 18:00-22:30
Investigators: Nadja ,Niels ,Jessica
Materials
Sequencing by eurofins mwg|operon
Label - Barcode
- 1cB - AKM001W053
- 1cC - AKM001W054
- 1cA - AKM001W055
- 2cB - AKM001W056
- 2cA - AKM001W057
- 2cD - AKM001W058
- 2cC - AKM001W059
- 1cD - AKM001W060
sequencing of
Time: 2011-08-23, 18:00-22:30
Investigators: Nadja ,Niels ,Jessica
Materials
Sequencing by eurofins mwg|operon
Label - Barcode
- K3CAcA - AKM001W061
- A3CAcA - AKM001W062
- A3YAcA - AKM001W063
- K3YIcC - AKM001W064
- K3CIcB - AKM001W065
- A3YIcB - AKM001W066
- A3cIcA - AKM001W067
over night culture
Time: 2011-08-23, 18:00-22:30
Investigators: Niels, Jessica, Nadja
Materials
- 5 ml LB Media
- 5 µl Amp/Kan
- clones
- pSBA3-YFP+const
- pSBA3-CFP+const A
- pSBA3-CFP+const C
- pSBK3-YFP+const A
- pSBK3-CFP+const A
- pSBK3-YFP+ara V
overnight 37°C
further tasks:
- miniprep
74th Labday 2011-08-24
Transformation w/ pUP089 A and B, BBa_K40304_Affibody_MiddleLinker+Ara/Const/Lac, pSB1T3+YFPI+Ara/Const/Lac and pSB1T3+YFPII+Ara/Const/Lac
Investigators: Nicole, Nadine
Time: 7:00-9:00
Aim:
- creating pARW089 without Amp resistance, called then pUP089
- get expression backbones w/ Tet or Cm resistance and Ara, Const. or Lac promotors
Material:
- XL1-blue (competent)
- RV-308 (competent)
- ligation products from o.n. ligation from 2011-08-23 (Jessica)
- pSB1C3 + Ara
- pSB1C3 + Lac
- pSB1C3 + const
- pSB1C3 + H2O
- pSB1T3-YFP II + Ara
- pSB1T3-YFP II + Lac
- pSB1T3-YFP II + const
- pSB1T3-YFP II + H2O
- pSB1T3-YFP I + Ara
- pSB1T3-YFP I + Lac
- pSB1T3-YFP I + const
- pSB1T3-YFP I + H2O
- pSB1T3-YFP I + H2O
- pARW089 AvaII A = pUP089 A
- pARW089 AvaII B = pUP089 B
- for pSB1T3 plasmids RV-308 cells were used
- for others: Xl1-blue
- LB-Agar
- tetracycline
- chloramphenicol
- kanamycine
Method:
- addition of 2 µl ligation reaction to XL1-blue cells
- incubation 60 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates:
- pUP089
- 100 µg/ml ampicilin
- 100 µg/ml ampicilin
- 100 µg/ml kanamycin
- 100 µg/ml kanamycin
- 100 µg/ml ampicilin and kanamycin
- 100 µg/ml ampicilin and kanamycin
- pSB1T3
- 100 µg/ml tetracyclin
- pSB1C3
- 100 µg/ml chloramphenicol
- incubation for 20 hrs at 37°C
Further tasks:
control cell clones
Miniprep of E.coli overnight culture containing created pPDV089
Investigators: Sabine, Laura
Aim: purification of pPDV089 for test digestion and sequencing
Time: 10:00-15:00
Method/Materials:
- see protocol 5.1 of the NucleoSpin Plasmid Kit
- 5 clones containing pSB1C3+mdnA
- 5 clones containing pSB1C3-geneIII
- 5 clones containing pSB1C3-mdnA/geneIII
- 10 clones containing pARW089+mdnA
- 10 clones containing pARW089+mdnA/geneIII (3 fragment ligation)
- 10 clones containing pARW089+mdnA/geneIII (2 step ligation)
Further tasks: test digestion
Test digestion of purified plasmids pARW089
Investigators: Sabine
Aim:control of liagation of mdnA and geneIII into pARW089
Time: 13:00-17:00
Materials/Methods:
- 0,5 µl XbaI
- 0,5 µl SpeI
- 2 µl 10x buffer 4 (NEB)
- 0,2 µl BSA
- 2 µl vector DNA (ca. 1 µg)
- 14,8 µl water
- incubate for 3 h at 37°C
Results:
- expected band sizes for pARW089 containing mdnA and geneIII: ca. 5300 bp, 4500 bp, 630 bp and 90
- expected band sizes for pARW089 not containing the inserts: ca. 10000 bp, 630 bp and 90 bp
Test digestion of purified plasmids pSB1C3
Investigators: Sabine
Aim:control of liagation of geneIII, mdnA and mdnA/geneIII into pSB1C3
Time: 18:00-18:30
Materials/Methods:
- per reaction:
- 0,5 µl XbaI
- 0,5 µl PvuI
- 2 µl 10x buffer 3 (NEB)
- 0,2 µl BSA
- 5 µl vector DNA (ca. 1 µg)
- 11,8 µl water
- incubate over night at 37°C
Results:
Agarose gel electrophoresis of digested mdn fragments and pSB1C3, gel extraction and ligation
Investigators: Nicole, Jessica, Steffi, Niels
Time: 2011-08-24,
Materials
- digests of :
- mdnB
- mdnC
- mdnD
- mdnE
- mdnBC
- mdnDE
- mdnABC
- pSB1C3
- agarose, 1x TAE, gel red
- gel extraction kit (Promega)
- thermosensitive alkaline phoshatase (TSAP, Promega)
- ligase
Gel electrophoresis:
- 1,25% agarose gel (0.625 g agarose + 50 ml 1x TAE, 2 µl gel red)
- 30 µl digest + 6 µl 6x loading dye
Gel 1
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | mdnB | 15 | ~1021 |
| 2 | mdnB | 15 | ~1021 |
| 3 | mdnC | 15 | ~1007 |
| 4 | mdnC | 15 | ~1007 |
| 5 | mdnD | 15 | ~600 |
| 6 | mdnD | 15 | ~600 |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 10 |
Gel 2
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | mdnE | 36 | ~2100 |
| 2 | mdnBC | 36 | ~2045 |
| 3 | mdnDE | 36 | ~2800 |
| 4 | mdnABC | 36 | ~2492 |
| 5 | - | ||
| 6 | pSB1C3 | 36 | ~2389, 243 |
Gel extraction:
- using Promega- Wizard SV Gel and PCR Clean Up System
Dephosphorylation of pSB1C3:
- using Promega TSAP
- adding 5 µl Buffer and 1 µl TSAP
- incubate at 37°C for 15 min
- heat inactivaton at 74°C for 15 min
- heat inactivaton at 74°C for 15 min
Nanodrop
| sample | c [ng/µl] | 260nm/280nm | 260nm/230nm |
| mdnB | 10,2 | 2,08 | 0,57 |
| mdnC | 13,4 | 2,3 | 0,4 |
| mdnD | 11,8 | 1,94 | 0,4 |
| mdnE | 13,3 | 1,93 | 0,78 |
| mdnBC | 16,3 | 1,91 | 0,79 |
| mdnDE | 13,8 | 1,92 | 0,60 |
| mdnABC | 13,4 | 1,89 | 0,68 |
| C3 | 44,4 | 1,90 | 1,45 |
Ligation:
| Insert | [µl] | Vector | [µl] |
| mdnB | 6 | C3 | 2 |
| mdnC | 6 | C3 | 2 |
| mdnD | 7 | C3 | 1 |
| mdnE | 5 | C3 | 3 |
| mdnBC | 5 | C3 | 3 |
| mdnDE | 5 | C3 | 3 |
| mdnABC | 5 | C3 | 3 |
- 1 µl 10x Buffer
- 1 µl T4 Ligase
- over night by 14°C
further tasks:
- transformation
Digest of pSB1A3+CFP/YFP and pSB1K3+CFP/YFP vectors w/ promotors
Investigators: Jessica, Nadja
Time: 2011-08-24, 14:00-20:00
Materials
- vectors from 2011-08-24:
- pSB1A3-CFP + const (clone A)
- pSB1A3-CFP + const (clone B)
- pSB1A3-CFP + const (clone C)
- pSB1A3-YFP + const (clone C)
- pSB1K3-YFP +Ara (clone V)
- pSB1K3-YFP +Ara (clone V box)
- pSB1K3-YFP +Ara (clone VI)
- pSB1K3-CFP + const (clone A)
- pSB1K3-CFP +const (clone B)
- pSB1K3-CFP +const (clone C)
- pSB1K3-YFP + const (clone A)
- pSB1K3-YFP +const (clone B)
- pSB1K3-YFP + const (clone C)
- pSB1K3-YFP + const (clone C)
- NdeI
- NruI
- HincII
- NotI
- BSA (100x)
- Buffer 3
- water
Digest:
- Reaction mix (Arabinose)
- 0.5 µl DNA
- 0.5 µl NdeI
- 0.5 µl NruI
- 2 µl Buffer 3 (10x)
- 16.5 µl water
- Reaction mix (constitutive)
- 0.5 µl DNA
- 0.5 µl HincII
- 0.5 µl NotI
- 0.2 µl BSA
- 2 µl Buffer 3 (10x)
- 16.3 µl water
- total volume each: 20 µl
- incubation at 37°C for 1.5 hrs
Gel electrophoresis:
- 1% agarose gel (0.5 g agarose + 50 ml 1x TAE, 2 µl gel red)
- 20 µl digest + 4 µl 6x loading dye
Gel 1
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 20 | |
| 1 | pSB1A3-CFP + const (clone A) (undigested) | 6 | |
| 2 | pSB1A3-CFP + const (clone A) | 15 | 2145, 804 |
| 3 | pSB1A3-CFP + const (clone B) | 15 | 2145, 804 |
| 4 | pSB1A3-CFP + const (clone C) | 15 | 2145, 804 |
| 5 | pSB1A3-YFP + const (clone C) | 15 | 2145, 804 |
| 6 | - | ||
| 7 | pSB1K3-YFP +Ara (clone V) undigested | 6 | |
| 8 | pSB1K3-YFP +Ara (clone V) | 15 | ~ 2800, 870, 430 |
| 9 | pSB1K3-YFP +Ara (clone V box) | 15 | ~ 2800, 870, 430 |
| 10 | pSB1K3-YFP +Ara (clone VI) | 15 | ~ 2800, 870, 430 |
Gel 2
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 20 | |
| 1 | - | ||
| 2 | pSB1K3-CFP + const (clone A) | 15 | 2145, 804 |
| 3 | pSB1K3-CFP +const (clone B) | 15 | 2145, 804 |
| 4 | pSB1K3-CFP +const (clone C) | 15 | 2145, 804 |
| 5 | pSB1K3-YFP + const (clone A) | 15 | 2145, 804 |
| 6 | pSB1K3-YFP +const (clone B) | 15 | 2145, 804 |
| 7 | pSB1K3-YFP + const (clone C) | 15 | 2145, 804 |
| 8 | pSB1K3-CFP + const (clone A) undigested | 6 |
further tasks:
- repeat digest of pSB1K3 + const with more DNA
- have a look at digest of pSB1K3 + Ara
Transformation w/ pUP089 A and B, BBa_K40304_Affibody_MiddleLinker+Ara/Const/Lac, pSB1T3+YFPI+Ara/Const/Lac and pSB1T3+YFPII+Ara/Const/Lac
Investigators: Nicole, Nadine
Time: 7:00-9:00
Aim:
- creating pARW089 without Amp resistance, called then pUP089
- get expression backbones w/ Tet or Cm resistance and Ara, Const. or Lac promotors
Material:
- XL1-blue (competent)
- RV-308 (competent)
- ligation products from o.n. ligation from 2011-08-23 (Jessica)
- pSB1C3 + Ara
- pSB1C3 + Lac
- pSB1C3 + const
- pSB1C3 + H2O
- pSB1T3-YFP II + Ara
- pSB1T3-YFP II + Lac
- pSB1T3-YFP II + const
- pSB1T3-YFP II + H2O
- pSB1T3-YFP I + Ara
- pSB1T3-YFP I + Lac
- pSB1T3-YFP I + const
- pSB1T3-YFP I + H2O
- pSB1T3-YFP I + H2O
- pARW089 AvaII A = pUP089 A
- pARW089 AvaII B = pUP089 B
- for pSB1T3 plasmids RV-308 cells were used
- for others: Xl1-blue
- LB-Agar
- tetracycline
- chloramphenicol
- kanamycine
Method:
- addition of 2 µl ligation reaction to XL1-blue cells
- incubation 60 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates:
- pUP089
- 100 µg/ml ampicilin
- 100 µg/ml ampicilin
- 100 µg/ml kanamycin
- 100 µg/ml kanamycin
- 100 µg/ml ampicilin and kanamycin
- 100 µg/ml ampicilin and kanamycin
- pSB1T3
- 100 µg/ml tetracyclin
- pSB1C3
- 100 µg/ml chloramphenicol
- incubation for 20 hrs at 37°C
Further tasks:
control cell clones
Miniprep of o. n. culture of pSB1K3 constitutive and Ara as well as pSB1A3 constitutive and Ara
Time: 7:00-
Investigators: Nicole, Nadine, Nadja
Results:
Concentration:
- pSBA3-YFP+const C = 470,8 ng/µl
- pSBA3-CFP+const A = 889,2 ng/µl
- pSBA3-CFP+const C = 692,8 ng/µl
- pSBK3-YFP+const A = 495,0 ng/µl
- pSBK3-CFP+const A = 1142,0 ng/µl
- pSBK3-YFP+ara V = 380,1 ng/µl
Further task:
- sequencing
75th Labday 2011-08-25
check plates from transformation (expression backbones in XL1-blue, 2011-08-24)
Time: 7:00-7:20
Investigators: Nicole, Nadine
Aim:
- build expression backbones w/ different promotors
Material:
- on chloramphenicol plates (in XL1 blue)
- pSB1C3 + Ara
- pSB1C3 + Lac
- pSB1C3 + const
- pSB1C3 + H2O
- pSB1T3-YFP II + Ara
- pSB1T3-YFP II + Lac
- pSB1T3-YFP II + const
- pSB1T3-YFP II + H2O
- pSB1T3-YFP I + Ara
- pSB1T3-YFP I + Lac
- pSB1T3-YFP I + const
- pSB1T3-YFP I + H2O
- pSB1T3-YFP I + H2O
Results:
- buffer control was over grown
conclusions:
- something is wrong with competent cells
Further tasks:
- check competent cells
check plates from transformation (pUP089 in XL1-blue, 2011-08-24)
Time: 7:00-7:20
Investigators: Nicole, Nadine
Aim:
- creating pARW089 without Amp resistance, called then pUP089
Material:
- pUP089 A in XL1 blue
- pUP089 B in XL1 blue
- on kan, amp or kan and amp plates
Results:
conclusions:
- elimination of ampicillin resistance has worked
- pUP089 vector will be used for mdnA library
Sequencing of pSB1K3 constitutive and Ara as well as pSB1A3 constitutive and Ara from 2011-08-24
Time: 19:00-
Investigators: Jessica, Nadja
Materials:
- pSBA3-YFP+const C = 470,8 ng/µl
- pSBA3-CFP+const A = 889,2 ng/µl
- pSBA3-CFP+const C = 692,8 ng/µl
- pSBK3-YFP+const A = 495,0 ng/µl
- pSBK3-CFP+const A = 1142,0 ng/µl
- pSBK3-YFP+ara V = 380,1 ng/µl
- Primer : VF2
- eurofins mwg/ operon Label-Barcodes
Method:
- 70µg/µl DNA, total volume of 15µl
- 2 pmol/µl Primer, ca. 10µl for each sample
Further task:
- analysing results
Transformation of pSB1C3 with several mdn genes/ promotor and pSB1T3-YFP I/II with several promotor
Investigators: Niels, Jessica
Material:
- XL1-blue (competent)
- RV-308 (competent)
- ligation products :
- pSB1C3 + mdnB
- pSB1C3 + mdnC
- pSB1C3 + mdnD
- pSB1C3 + mdnE
- pSB1C3 + mdnBC
- pSB1C3 + mdnABC
- pSB1C3 + mdnDE
- pSB1C3 + Ara
- pSB1C3 + Lac
- pSB1C3 + const
- pSB1C3 + H2O
- pSB1T3-YFP II + Ara
- pSB1T3-YFP II + Lac
- pSB1T3-YFP II + const
- pSB1T3-YFP II + H2O
- pSB1T3-YFP I + Ara
- pSB1T3-YFP I + Lac
- pSB1T3-YFP I + const
- pSB1T3-YFP I + H2O
- pSB1T3-YFP I + H2O
- for pSB1T3 plasmids RV-308 cells were used
- for others: Xl1-blue
- LB-Agar
- tetracycline
- chloramphenicol
Method:
- addition of 2 µl ligation reaction to XL1-blue cells
- incubation 60 min on ice,
- heat shock 90 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates:
- pSB1T3
- 100 µg/ml tetracyclin
- pSB1C3
- 100 µg/ml chloramphenicol
- incubation over night at 37°C
Further tasks:
control cell clones
Results (from 2011-08-26):
- for transformations of pSB1C3 + mdn fragments 2 clones were picked from each plate
- plates of pSB1C3 and pSB1T3 with promoters were thrown away because of too many colonies on control
PCR of complete mdn cluster for biobrick-construction
Time: 2011-08-25
Investigators: Nadine, Jessica
Material:
- pARW089, 20.8.11, 3.3 ng/µl
- pARW071, 27.5.11, 6.3 ng/µl
- dNTPs
- primer 77, 78, 79, 80
- HF Phusion Buffer 5x* Phusion Polymerase
Method:
- 2µl pARW089, 1µl pARW071, respectively
- 1µl dNTPs
- 2µl forward primer (10mM)
- 2µl reverse primer (10mM)
- 10µl HF Phusion Buffer 5x
- 0.5 µl Phusion Polymerase
- 31.5 µl water (total volume= 50µl)
- reaction
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 95°C | 30 s |
| Annealing | 59°C | 60 s |
| Extension | 72°C | 2 min |
| DenaturationII | 95°C | 30 s |
| AnnealingII | 59°C | 60 s |
| ExtensionII | 72°C | 2 min |
| Final extension | 68°C | 5 min |
Result:
- three different PCR samples
- PCR sample 1: mdnABCDE from pARW071, Primer: 77/78
- PCR sample 2: mdnABCDE with T7 from pARW089, Primer: 77/79
- PCR sample 3: mdnABCDE without T7 from pARW089, Primer: 77/80
Output:
- mdnABCDE71
- mdnABCDE89-T7
- mdnABCDE89
Further Tasks:
- agarose gel
- PCR clean up
- restriction enzyme digestion
- ligation
- transformation
gel purification of AraC, TEV, 14_3C and TEv vector and 14_3C vector
Investigator: Stefan
Material:
- Gel purification kit (Machery & Nagel)
Methode:
protocol for gel purification.
Further tasks:
ligation of different fragmentsand transformation of competent E.coli XL1 blue cells.
ligation of AraC, TEV/14_3C with the TEV/14_3C vector
Investigator: Stefan
Material:
| Column heading 1 | TEV 1 µL | TEV 2 µL | TEV 3 µL | TEV 4 µL | 14_3C 1 µL | 14_3C 2 µL | control 14_3C vector µL | control TEV vector µL | |
|---|---|---|---|---|---|---|---|---|---|
| AraC | 0.8 (35 ng) | 1.8 (13.5 ng) | 1.8 (8.8 ng) | 2.3 (8.8 ng) | 3.4 (7.5 ng) | 1.4 (35 ng) | 0 | 0 | |
| Protease | 1.4 (11.9 ng) | 1.3 (6.3 ng) | 0.3 (61.9 ng) | 0.7 (20.2 ng) | 1.8 (7.1 ng) | 3.6 (61.9 ng) | 0 | 0 | |
| vector | 5.8 (6.4 ng) | 4.9 (6.4 ng) | 6 (5.2 ng) | 5.1 (5.2 ng) | 2.8 (11.6 ng) | 3. (22.2 ng) | 5.8 (6.4 ng) | 4.9 (6.4 ng) | |
| buffer | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| ligase | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| H2O | 0 | 0 | 0 | 0 | 0 | 0 | 2.8 | 5.2 |
Method:
ligation at room temperatur for 1 h
Further tasks:
transformation in competent cells
transformation of ligations
Investigator: Stefan
Material:
Method:
streak out of LB plattes with CM, use 150 µL and spin down the rest and streak it out, too.
Further tasks:
checking clones
over night culture of pUP089 A/*B from 24.08.2011
Investigators: Niels, Nadja
Materials/Method:
- 5 ml LB media
- 5 µl Kanamycin (50mg/ml)
- clone of
- pUP089 A - I
- pUP089 A - II
- pUP089 A - III
- pUP089 B - I
- pUP089 B - II
- pUP089 B - III
- 37°C, 250 rpm, over night
Further task:
- miniprep
PCR of geneIII and mdnA
Investigators: Sabine
Aim:
- amplification of mdnA and geneIII for cloning into pSB1C3
Reaction Components:
- 2 µl/ 20 ng geneIII (cut out from pARW089)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl primer pf_geneIII_EheI_XbaI_NgoMIV
- 1 µl primer pr_geneIII_iGEM_AatII
- 5 µl 10x PCR Buffer S
- 39,75 µl water
- 5 µl/ 50 ng pak bla KDIR (mdnA)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl primer pf_mdnA_iGEM_EheI
- 1 µl primer pr_mdnA_iGEM_AatII
- 5 µl 10x PCR Buffer S
- 36,75 µl water
Further tasks:
- analytic gel electrophoresis
- purification
- digestion
Analytic gel electrophoresis to control PCR
Investigators: Sabine
Time: 12:00-13:00
Aim: control of PCR
Material/Method:
- PCR products (mdnA and geneIII)
- 1 % agarose gel
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (1:10) (Fermentas)
- 6x Loading Dye (Fermentas)
- Gel Red
Result:
- PCR product with right band sizes
- mdnA ca. 200 bp
- geneIII ca. 500 bp
Further tasks:
- purification
- digestion
prepare samples for sequencing (created pPDV089)
Investigators: Sabine
Time: 12:00-13:00
Aim: sequencing (GATC)
Material/Method:
- purified plasmids of clones 3F6, 3F8, 2S7 and 2S14 (80 ng/µl, 20 µl total volume)
- primer mdnA_6 (10 pmol/µl, 30 µl total volume)
Further tasks: control sequence
digestion of created pPDV089 for deletion of kanamycin gene
Investigators: Laura, Sabine
Aim:
- inactivation of kanamycin resistence gene
- reason: helper plasmid contains also kanamycin restistence gene
- enable selection of E. coli cells containing both helper plasmid and phage display vector
Time: 14:00-14:30
Reaction components:
- 3 µl pPDV089 (from clones 3F6, 3F8, 2S7 and 2S14)
- 1 µl restriction enzyme NsiI (Fermentas, AG Dittmann)
- 2 µl buffer (Fermentas, AG Dittmann)
- 14 µl water
Reaction conditions:
- over night at 37°C
Digestion of PCR products geneIII and mdnA for cloning into pSB1C3
Investigator: Laura, Sabine
Aim: cloning of mdnA, geneIII and mdnA/geneIII into pSB1C3
Time: 14:30-16:00
Material/Method:
- 30 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 4 µl NEB 10x buffer 2
- 0,4 µl BSA
- 3,6 µl water
- 1,5 h, 37°C
- 20 µl PCR product geneIII
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 3 µl NEB 10x buffer 2
- 4,7 µl water
- 0,3 µl BSA
- 1,5 h, 37°C
- 20 µl PCR product geneIII
- 1 µl restriction enzyme NgoMIV
- 1 µl restriction enzyme PstI
- 3 µl NEB 10x buffer 1
- 0,3 µl BSA
- 4,7 µl water
- 1,5 h, 37°C
- 20 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme AgeI
- 3 µl NEB 10x buffer 1
- 4,7 µl water
- 0,3 µl BSA
- 1,5 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation of mdnA and geneIII into pSB1C3
Agarose gel electrophoresis for purification of digested PCR products
Investigator: Sabine
Aim: control and purification of digested PCR products mdnA and geneIII
Time: 16:30-17:30
Material/Method:
- PCR products (mdnA and geneIII)
- 1 % agarose gel
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (1:10) (Fermentas)
- 6x Loading Dye (Fermentas)
- Gel Red
76th Labday 2011-08-26
miniprep of pUP089 clones
Investigators: Nadine
Time: 2011-08.26, 8:00-9:30
Aim:
- glycerol stocks
- purification of pUP089 plasmids for mdnA library (and screening)
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl Elution buffer
- Check concentration with NanoDrop
- o.n. cultures:
- pUP089 A I
- pUP089 A II
- pUP089 A III
- pUP089 B I
- pUP089 B II
- pUP089 B III
- pUP089 B III
Results:
- pUP089 A I : 498.5 ng/µl
- pUP089 A II: 465.2 ng/µl
- pUP089 A III: 462.2 ng/µl
- pUP089 B I: 426.1 ng/µl
- pUP089 B II: 443.1 ng/µl
- pUP089 B III: 448.9 ng/µl
Output:
- # P15: pUP089 A1
- # P16: pUP089 A II
- # P17: pUP089 A III
- # P18: pUP089 B I
- # P19: pUP089 B II
- # P20: pUP089 B III
- glycerol stocks in XL1 blue:
- # G2 pUP089 A I
- # G3 pUP089 A II
- # G4 pUP089 A III
- # G5 pUP089 B I
- # G6 pUP089 B II
- # G7 pUP089 B III
PCR of complete mdn cluster for biobrick-construction
Investigators: Steffi, Nadine, Katharina
Material:
- pARW089
- pARW071
- dNTPs
- primer 77, 78, 79, 80
- HF Phusion Buffer 5x* Phusion Polymerase
Method:
- 2µl pARW089, 1µl pARW071, respectively
- 1µl dNTPs
- 2µl forward primer (10mM)
- 2µl reverse primer (10mM)
- 10µl HF Phusion Buffer 5x
- 0.5 µl Phusion Polymerase
- 31.5 µl water (total volume= 50µl)
- reaction
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 95°C | Hold |
| Initial denaturation | 95°C | 30 sec |
| Denaturation | 95°C | 30 s |
| Annealing | 51°C | 60 s |
| Extension | 68°C | 6.5 min |
| DenaturationII | 95°C | 30 s |
| AnnealingII | 63°C | 60 s |
| ExtensionII | 68°C | 6.5 min |
| Final extension | 68°C | 50 min |
Result:
- three different PCR samples
- PCR sample 1: pARW071 + primer 77/78 = mdnABCDE71
- PCR sample 2: pARW089 + primer 77/79 = mdnABCDE89+T7
- PCR sample 3: pARW089 + primer 77/80 = mdnABCDE89
Further Tasks:
- gel electrophoresis
- PCR clean up
- restriction enzyme digestion
- ligation
- transformation
digest of PCR products from 25.08.2011
Investigators: Niels
Aim:
- digest of PCR - products
Materials: PCR from 25.08.2011
- PCR loop
- PCR 1 Libary
- PCR 2 Libary
Sample:
- 0,5 µl DNA
- 1 µl Aat II (Roche)
- 3 µl 10x Buffer (Roche)
- 25,5 µl water
- total volume 30 µl
- 37°C
- 3 hours
further tasks:
- ligation with pUP089
Repeated ligation of digested mdn fragments and pSB1C3 ( from 2011-08-24)
Investigators: Jessica, Nadja
Time: 2011-08-26, 18:00
Materials
Ligation:
| Insert | [µl] | Vector | [µl] |
| mdnB | 6 | C3 | 2 |
| mdnC | 6 | C3 | 2 |
| mdnD | 7 | C3 | 1 |
| mdnE | 5 | C3 | 3 |
| mdnBC | 5 | C3 | 3 |
| mdnDE | 5 | C3 | 3 |
| mdnABC | 5 | C3 | 3 |
| H20 | 5 | C3 | 3 |
- 1 µl 10x Buffer
- 1 µl T4 Ligase
- over night by 14°C
Further tasks:
- transformation
Agarose gel and Gel extraction of pUP089 and the three libraries as well as their ligation
Investigators: Jessica, Nadine, Nadja
Time: 2011-08-26, 16:00
Aim
- Build up a Library of mutated mdnA`s
Materials
- digests of:
- pUP089
- libraries 800, 2000, loop
Agarose gel
Gel 1
- 1%: 0.5 g agarose + 50 ml 1x TAE, adding 2 µl gel red
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 12 | |
| 1 | - | ||
| 2 | digest pUP089 | 36 | 10029, 161 |
Gel 2
- 2%: 1 g agarose + 50 ml 1x TAE, adding 2 µl gel red
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ 100bp DNA Ladder | 2 | |
| 1 | |||
| 2 | Library 1 | 36 | 161 |
| 3 | |||
| 4 | Library 2 | 36 | 161 |
| 5 | - | ||
| 6 | Library Loop | 36 | 161 |
Gel extraction
- Promega- Wizard SV Gel and PCR Clean Up System
- Eluation with 50µl Nuclease- Free Water
Nano- Drop :Concentrations
- Library 1- digested- purified 11-26: 6,2 ng/µl
- Library 2- digested- purified 11-26: 6,1 ng/µl
- Library Loop- digested- purified 11-26: 7,5 ng/µl
- pUP089: 11,3 ng/µl
Dephosphorylation of pUP089
- add 5µl Multicore 10x Buffer (Promega)
- add 1µl Thermosensitive Alkaline Phosphatase (Promega)
- incubate at 37°C for 15 min
- heat inactivation at 74°C for 15 min
Ligation
| Insert | [µl] | Vector | [µl] |
| Library 1 | 1 | pUP089 | 7 |
| Library 2 | 1 | pUP089 | 7 |
| Library Loop | 1 | pUP089 | 7 |
| H2O | 1 | pUP089 | 7 |
- 1 µl 10x Buffer
- 1 µl T4 Ligase
- over night at 14°C
Further tasks:
- transformation
over night cultures of XL1 blue pSB1C3+mdn-combination
Time: 2011-08-26, 10:00-12:00
Investigators: Nadine, Nadja
Material:
- plates from 2011-08-25, Niels
- pSB1C3+mdnB clone 1
- pSB1C3+mdnB clone 2
- pSB1C3+mdnC clone 1
- pSB1C3+mdnCclone 2
- pSB1C3+mdnD clone 1
- pSB1C3+mdnD clone 2
- pSB1C3+mdnE clone 1
- pSB1C3+mdnE clone 2
- pSB1C3+mdnABC clone 1
- pSB1C3+mdnABC clone 2
- pSB1C3+mdnBC clone 1
- pSB1C3+mdnBC clone 2
- pSB1C3+mdnDE clone 1
- pSB1C3+mdnDE clone 2
- 25 mg/ml chloramphenicol
- LB-medium
Protocol:
- 5 ml LB and 5 µl chloramphenicol
- tip w/ colony
- incubation: 37°C, 200 rpm, over night
Glycerol stocks of E. coli containing created pPDV089
Investigator: Sabine
Time: 10:30-10:30
Material/Method:
- 300 µl glycerol per stock
- 700 µl cell culture per stock
- storage at 80 °C
Gel electrophoresis to control digestion of pPDV089 with NsiI
Investigators: Sabine
Time: 10:30-12:00
Aim: control of digestion and purification
Material/Method:
- digested pPDV089 (clones 3F6, 3F8, 2S7 and 2S14)
- 1 % agarose gel
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (1:10) (Fermentas)
- 6x Loading Dye (Fermentas)
- Gel Red
- purification see protocol NucleoSpin ExtractIII kit
Result:
- right band sizes
- vector ca. 10 kb
- 2 bands closed to each other near 10 kb (except 3F8), both were separately cut out of the gel)
- deleted part of kanamycin gene ca. 300 bp
Further tasks:
- purification
- re-ligation
PCR of geneIII
Investigators: Sabine
Time: 12:00-14:00
Aim: amplification of geneIII for cloning into pARW089
Reaction Components:
- 2 µl/ 20 ng geneIII (cut out from pARW089)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl primer pf_geneIII_EheI_XbaI_NgoMIV
- 1 µl primer pr_geneIII_iGEM_AatII
- 5 µl 10x PCR Buffer S
- 39,75 µl DNase free water
- prufication see protocol NucleoSpin ExtractII kit
Further tasks:
- digestion
Ligation go geneIII and mdnA into pSB1C3
Investigator: Sabine
Aim: cloning of mdnA, geneIII and mdnA/geneIII into pSB1C3
Time: 14:00-15:30
Material/Method:
- 1,9 µl PCR product geneIII (36 µg/µl)
- 6,1 µl pSB1C3 (17 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 1 h at room temperature
- 2,4 µl PCR product mdnA (12 µg/µl)
- 6,6 µl pSB1C3 (17 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 1 h at room temperature
- 1,9 µl PCR product mdnA (12 µg/µl)
- 1,6 µl PCR product geneIII (36 µg/µl)
- 4,5 µl pSB1C3 (17 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Re-ligation of NsiI-digested pPDVs089
Investigator: Sabine
Aim: create pPDV089 without kanamycin resistence
Time: 14:30-16:00
Material/Method:
- 17 µl NsiI-digested pPDVs089 (3F6, 3F8, 2S7 and 2S14, DNA from upper and lower bands)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Ligation of geneIII into pARW089
Investigator: Sabine
Aim: cloning of geneIII into pARW089
Time: 15:00-17:30
Material/Method:
- 1,5 µl PCR product geneIII (17 µg/µl)
- 6,5 µl pARW089 (30 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Transformation of created vectors in E. coli
Investigators: Sabine
Aim:amplification of vectors
Time: 18:00-21:00
Material:
- pPDV089 without kanamycin resistence (3F6, 3F8, 2S7 and 2S14)
- pARW089 containing only geneIII (no mdnA)
- pSB1C3 containing mdnA, geneIII or mdnA/geneIII
- agar plates containing chloramphenicol (pSB1C3)
- agar plates containing kanamycin, ampicillin and kanamycin+ampicillin (pPDV)
- agar plates containing kanamycin (pARW089)
Method:
- addition of 4 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
control cell clones
77th Labday 2011-08-27
Agarose gel of PCR prducts mdnABC and PCR purification
Time: 2011-08-27, 10:00-12:00
Investigators: Nadine
Aim:
- prove PCR
- purification of PCR prducts
Materials:
- PCR products from 2011-08-26 , Katharina
- mdnABCDE71
- mdnABCDE89+T7
- mdnABCDE89
- Promega- Wizard SV Gel and PCR Clean Up System (elution in 50 µl elution buffer)
- Agarose (BioRad)
- 1x TAE
- gel red
- loading dye (promega)
- 1:100 ladder mix (Fermentas)
Protocol:
- 1%: 0.5 ge agaros + 50 ml 1xTAE buffer, 2 µl gel red
- PCR clean up: following protocol from kit
Results:
| lane | Sample | Volume in µl | Expected size in bp |
| M | Ladder | 12 | |
| 1 | |||
| 2 | mdnABCDE from pARW071 | 12 (2 µl PCR product) | ~6500 |
| 3 | |||
| 4 | mdnABCDE from pARW089 w/ T7 | 12 (2 µl PCR product) | ~6500 |
| 5 | - | ||
| 6 | mdnABCDE from pARW089 without T7 | 12 (2 µl PCR product) | ~6500 |
- no product for mdnABCDE of pARW089 w/ T7
- PCR clean up:
- mdnABCDE from pARW071: 36.6 ng/µl
- mdnABCDE from pARW089 w/ T7: 37.6 ng/µl
- mdnABCDE from pARW089 without T7: 63.9 ng/µl
Further tasks:
- digest w/ EcoRI and SpeI (also pSB1C3)
- repeat PCR (third time)
competent cells - E.coli XL1 blue & RF308
Investigator: Steffi
Aim: produce competent cells
Materials/Methods:
| TFB I | 1000ml | 200ml |
| 100mM Rubidium Chloride | 12.1 | 2.42g |
| 30mM Potassium Acetate | 2.944 | 0.59g |
| 10mM Calcium Chloride | 1.47 | 0.29g |
| 15% w/v Glycerol (87%) | 150 | 34.5g |
Adjust pH to 5.8 with acetic acid
Filter sterilize the solution
| TFB II | 500ml | 100ml |
| 50mM Rubidium Chloride | 0.6 | 0.121g |
| 10mM MOPS | 1.05 | 0.210g |
| 75mM Calcium Chloride | 5.51 | 1.100g |
| 15% w/v Glycerol (87%) | 75 | 17.24g |
Adjust pH to 7.0 with KOH
Filter sterilize the solution
Work always sterile and cold and speedy!
- All volumes deal with the common cellline!
- Prepare 68 Eppis (1,5µl)
- get liquid nitrogen
- prepare 5 ml LB-Medium with the specific antibiotic (for XL1-blue: Tet, for BL21: none!), inoculate and incubate over night
- prepare 200 ml LB-Medium with the specific antibiotic, inoculate with 2 ml of the over-night-culture
- grow while shaking at 37°C, 190 rpm to an OD600 at 0,4-0,6
- keep cell suspension in sterile falcons (50 ml) 10 min on ice, then centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend on ice with 40 ml icecold TFB I and keep 10 min on ice
- centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend pellet in 8 ml TFB II
- aliquot in Eppis: 50µl per tube and store immediately at liquid nitrogen and afterwards at -80 °C
Results:
- 34 tubes RV308 for expression(50 µl competent cells at -80°C)
- 34 tubes XL1 blue for transformation(50 µl competent cells at -80°C)
Further tasks:
check by transformation
miniprep of several mdn clones and glycerole stocks
Investigators: Nadine, Katharina
Time: 2011-08-27, 11:00-12:00
Aim:
- glycerol stocks
- purification of pSB1C3+mdnX plasmids for mdnX biobricks (and screening)
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl Elution buffer
- Check concentration with NanoDrop
- o.n. cultures (from 2011-08-26, Nad/Nad):
- pSB1C3+mdnB clone 1
- pSB1C3+mdnB clone 2
- pSB1C3+mdnC clone 1
- pSB1C3+mdnCclone 2
- pSB1C3+mdnD clone 1
- pSB1C3+mdnD clone 2
- pSB1C3+mdnE clone 1
- pSB1C3+mdnE clone 2
- pSB1C3+mdnABC clone 1
- pSB1C3+mdnABC clone 2
- pSB1C3+mdnBC clone 1
- pSB1C3+mdnBC clone 2
- pSB1C3+mdnDE clone 1
- pSB1C3+mdnDE clone 2
Results:
- pSB1C3+mdnB clone 1: 778.3 ng/µl
- pSB1C3+mdnB clone 2: 600.5 ng/µl
- pSB1C3+mdnC clone 1: 1031.7 ng/µl
- pSB1C3+mdnCclone 2: 843.3 ng/µl
- pSB1C3+mdnD clone 1: 482.8 ng/µl
- pSB1C3+mdnD clone 2: 541.1 ng/µl
- pSB1C3+mdnE clone 1: 734.4 ng/µl
- pSB1C3+mdnE clone 2: 768.2 ng/µl
- pSB1C3+mdnABC clone 1: 853.4 ng/µl
- pSB1C3+mdnABC clone 2: 690.3 ng/µl
- pSB1C3+mdnBC clone 1: 626.9 ng/µl
- pSB1C3+mdnBC clone 2: 764 ng/µl
- pSB1C3+mdnDE clone 1: 899.4 ng/µl
- pSB1C3+mdnDE clone 2: 525.6 ng/µl
Output:
- plasmids: see results (stored in -20°C: green box, red label: mdn biobricks)
- glycerol stocks in XL1 blue (stored in -80°C, glycerole box #2):
- pSB1C3+mdnB clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnB clone 2, 27.8.11, Nad/Kat
- pSB1C3+mdnC clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnCclone 2, 27.8.11, Nad/Kat
- pSB1C3+mdnD clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnD clone 2, 27.8.11, Nad/Kat
- pSB1C3+mdnE clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnE clone 2, 27.8.11, Nad/Kat
- pSB1C3+mdnABC clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnABC clone 2, 27.8.11, Nad/Kat
- pSB1C3+mdnBC clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnBC clone 2, 27.8.11, Nad/Kat
- pSB1C3+mdnDE clone 1, 27.8.11, Nad/Kat
- pSB1C3+mdnDE clone 2, 27.8.11, Nad/Kat
Further Tasks:
- test digest (today)
- sequencing (Monday, 2011-08-29)
Digestion of PCR products of complete mdn-cluster (mdnABCDE)
Investigators: Nadine, Katharina
Materials
- pSB1C3
- purified PCR products of mdn-cluster
- NEB Buffer 4
- EcoRI
- SpeI
- BSA
Method
- digestion of inserts (PCR products of mdn-cluster)
- 50 µl purified PCR product
- 1.5 µl EcoRI
- 1.5 µl SpeI
- 6 µl NEB Buffer 4
- 0.6 µl BSA
- 0.4 µl sterile water
- digestion of vector (pSB1C3 235.0 ng/µl)
- 3 µl NEB Buffer 4
- 4 µl pSB1C3
- 0.3 µl BSA
- 1.5 µl EcoRI
- 1.5 µl SpeI
- 19.7 µl sterile water
Agarose gel
- 0.8%: 0.4 g agarose + 50 ml 1x TAE, adding 2 µl gel red
| lane | Sample | Volume in µl | Expected size in bp |
| M | Ladder | 12 | |
| 1 | mdnABC from pARW071, digested | ?? | ~6500 |
| 2 | mdnABC from pARW089 w/ T7, digested | ?? | ~6500 |
| 3 | mdnABC from pARW089 without T7, digested | ?? | ~6500 |
| 4 | pSB1C3, digested | ~150, 2400 |
Conclusions
- only mdnABCDE89 has worked
Further tasks:
- repeat PCR for mdnABCDE71 and mdnABCDE89 and mdnABCDE89+T7
- Transformation of pSB1C3+mdnABCDE89 in XL1 blue
Transformation of ligation: pUP089 + Lib-1/2, pSB1c3 + several mdn
Investigator: Niels
Material:
- XL1-blue (competent)
- ligation products from ligation from 2011-08-26/25
old competent cells
- pSB1C3 + H2O
- pSB1C3 + mdnBC
- pSB1C3 + mdnB
- pUP089 + puB control
- pUP089 + Lib-loop
- pUP089 + Lib-1
- pUP089 + Lib-2
new competent cells
- pSB1C3 + mdnE
- pSB1C3 + H2O
- pSB1C3 + mdnD
- pSB1C3 + mdnABC
- pSB1C3 + mdnDE
- pSB1C3 + mdnC
- puP089 + Lib-1
- LB-Agar
- chloramphenicol
- kanamycine
Method:
- addition of 2 µl ligation reaction to XL1-blue cells
- incubation 60 min on ice,
- heat shock 90 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates:
- pUP089
- 100 µg/ml kanamycin
- 100 µg/ml kanamycin
- pSB1C3
- 100 µg/ml chloramphenicol
- incubation for 20 hrs at 37°C
Further tasks:
control cell clones
Ligation of 3x pUP089 + Lib-2
Investigators: Niels
Material:
- pUP089 (restricted with EheI/AatII) (Vector)
- Lib-2 (Insert)
Method:
- 7 µl pUP089
- 1 µl Insert
- 1 µl Ligase buffer
- 1 µl T4 Ligase
Further tasks:
Transformation
Ligation of mdnABCDE89 + pSB1C3
Investigators: Katharina
Material:
- pSB1C3 (Vector)
- mdnABCDE89 (Insert)
Method:
- ligation
- 7 µl purified PCR product of mdn-cluster
- 1 µl pSB1C3
- 1 µl Ligase buffer
- 1 µl T4 Ligase
- ligation control
- 7 µl sterile water
- 1 µl pSB1C3
- 1 µl Ligase buffer
- 1 µl T4 Ligase
Mini-Prep of the positiv clones from an overnight culture
Investigator: Stefan
Material:
- Mini-Prep Kit (machery & Nagel)
Method:
Standard protocol 5.1 for plasmid preparation. And do glycerol stock cultures of all them.
Further tasks:
Send plasmids for sequencing
test digest of pSB1C3+mdncombinations (several mdn clones/combinations)
Time: 2011-08-27,16:30-18:30
Investigators: Nadine
Aim: prove of Insert (mdn combinations)
Materials:
- pSB1C3+mdn-combinations (miniprep from today, see above, Nad/Kat)
- HindIII, PvuII, ClaI, AvaI, HpaI (NEB)
- Buffer 4 and 2 (NEB, 10x)
- BSA
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnC | 3385 | HindIII | 2 | 363, 3022 |
| mdnD | 2944 | PvuII | 4 | 220, 591, 2133 |
| mdnE | 4474 | ClaI | 4 + BSA | 769, 3705 |
| mdnABC | 4852 | HindIII, AvaI | 2 | 363, 836, 3653 |
| mdnBC | 4419 | HindIII, AvaI | 2 | 363, 836, 3220 |
| mdnDE | 5154 | HpaI | 4 | 1329, 3825 |
Digestion protocol (one enzyme)
- 1 µl DNA
- 0.5 µl enzyme (see table)
- 2 µl 10x buffer
- 16.5 µl pure water
Digestion protocol (two enzymes)
- 1 µl DNA
- 0.5 µl enzyme I
- 0.5 µl enzyme II
- 2 µl 10x buffer
- 16 µl pure water
Digestion protocol (two enzymes and BSA)
- 1 µl DNA
- 0.5 µl enzyme I
- 0.5 µl enzyme II
- 2 µl 10x buffer
- 0.2 µl BSA
- 15.8 µl pure water
- total: 20µl
- 37°C for over night
Conclusions:
- annotation of mdnB in pARW089 (genious) might be wrong
Further tasks:
- agarose gel
- check annotation of mdnB
Troubleshooting of transformed cells
Investigator: Stefan
Aim:
- check of vector is digested at all
Materials:
heat shock transformation
Method:
heat shock transformation
Output:
78th Labday 2011-08-28
comparison of "new" and "old" competent cells
Investigators: Katharina
Results:
check plates from transformation (pSB1C3 w/ mdn combinations in XL1 blue, 2011-08-27)
Time: 2011-08-28, 12:00-12:30
Investigators: Katharina, Nadine
Aim:
- build biobricks w/ mdn combinations
Material:
- plates
- scanner
Results:
conclusions:
- Trafo has worked
Further tasks:
- pick colonies from plates
- mini prep tomorrow
- test digest (see 2011-08-27)
- sequencing
Agarose gel of PCR prducts mdnABC and PCR purification
Time: 2011-08-27, 10:00-12:00
Investigators: Nadine, Katharina
Aim:
- prove insert of pSB1C3 with mdn-combinations (biobricks) (miniprep from 2011-08-27, Nad/Kat)
Materials:
- Agarose (BioRad)
- 1x TAE
- gel red
- loading dye (promega)
- 1:100 ladder mix (Fermentas)
- 1kb DNA ladder (Promega)
Protocol:
- 2 x 1%: 0.5 g agarose + 50 ml 1xTAE buffer, 2 µl gel red
Results:
- gel 1
| lane | sample | enzymes | volume in µl | Expected size in bp |
| M1 | 1:100 DNA ladder mix, Fermentas | 12 | ||
| 1 | - | - | - | - |
| 2 | mdnABC 1 | HindIII, AvaI | 24 | 363, 836, 3653 |
| 3 | mdnABC 2 | HindIII, AvaI | 24 | 363, 836, 3653 |
| 4 | - | - | - | - |
| 5 | mdnBC 1 | HindIII, AvaI | 24 | 363, 836, 3220 |
| 6 | mdnBC 2 | HindIII, AvaI | 24 | 363, 836, 3220 |
| 7 | - | - | - | - |
| 8 | mdnC 1 | HindIII | 24 | 363, 3022 |
| 9 | mdnC 2 | HindIII | 24 | 363, 3022 |
| 10 | - | - | - | - |
| M2 | 1kb DNA ladder, Promega | - | 6 | - |
- gel 2
| lane | sample | enzymes | volume in µl | Expected size in bp |
| M1 | 1:100 DNA ladder mix, Fermentas | 12 | ||
| 1 | - | - | - | - |
| 2 | mdnD 1 | PvuI | 24 | 220, 591, 2133 |
| 3 | mdnD 2 | PvuI | 24 | 220, 591, 2133 |
| 4 | - | - | - | - |
| 5 | mdnDE 1 | HpaI | 24 | 1329, 3825 |
| 6 | mdnDE 2 | HpaI | 24 | 1329, 3825 |
| 7 | - | - | - | - |
| 8 | mdnE 1 | ClaI | 24 | 769, 3705 |
| 9 | mdnE 2 | ClaI | 24 | 769, 3705 |
| 10 | - | - | - | - |
| M2 | 1kb DNA ladder, Promega | - | 6 | - |
Conclusions:
- only D1 (Gel 2, lane 2) seems to be okay
over night cultures of XL1 blue pSB1C3+mdn-combination
Time: 2011-08-26, 10:00-12:00
Investigators: Katharina
Material:
- plates from 2011-08-27, Niels
new competent cells
- pSB1C3 + mdnE
- pSB1C3 + mdnD
- pSB1C3 + mdnABC
- pSB1C3 + mdnDE
- pSB1C3 + mdnC
- each plate: 3 clones
- 25 mg/ml chloramphenicol
- LB-medium
Protocol:
- 5 ml LB and 5 µl chloramphenicol
- tip w/ colony
- incubation: 37°C, 200 rpm, over night
Output:
- 15 tubes in shaker:
- pSB1C3 + mdnE: 1, 2, 3
- pSB1C3 + mdnD: 1, 2, 3
- pSB1C3 + mdnABC: 1, 2, 3
- pSB1C3 + mdnDE: 1, 2, 3
- pSB1C3 + mdnC: 1, 2, 3
Further tasks:
- mini prep tomorrow
Production of TEV and 14_3C biobricks part 1
Aim: indtroduction of iGEM restriction sites to produced mutated TEV and 14_3C Fragments.
Primer TEV:
(1) f_TEV_ACCAGC, r_TEV_iGEM_Eco81l
(2) r_TEV_ACCAGC, f_TEV_iGEM
Primer 14_3C:
(1) f_14_3C_ACCAGC, r_14_3C_iGEM_Eco81l
(2) r_14_3C_ACCAGC, f_14_3C_iGEM
Methode:
PCR
- Template: 1µl (TEV or 14_3C <10ng)
- Nucleotides: 1µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 2µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer)
- 35,5µl of pure water
- 0,5µl TaqPol
Program:
- Denat: 3min 94°C
- 5x:
Denat: 30sec 94°C
Anneal:30sec 55°C
Extend:30sec 72°C
- 25x:
Denat: 30sec 55°C
Anneal:30sec 55°C
Extend:30sec 72°C
- Final Extend: 10min 72°C
Further Tasks:
PCR purification of PCR products and Annealing of produced fragments
Transformation of pSB1C3+mdnABCDE89 and pUP089+library2
Investigators: Katharina, Steffi
Aim: Transformation of Ligation
Time: 2011-08-28, 11:00-13:00
Materials:
- competent E. coli cells (XL1-Blue, 2011-08-27, Steffi)
- ligation products: pSB1C3+mdnABCDE89 (2011-08-27, Katharina), pUP089+Lib2 (2011-08-27, Niels)
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Cm, Kan)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Output:
- 10 plates w/ Kan: mdn-lib1
- 2 plates w/ Cm: pSB1C3+mdnABCDE89 (and control)
overnight culture of picked E. coli clones transformed with pPDV089 to control deletion of kanamycin gene
Investigators: Sabine
Aim: control deletion of kanamycin gene in cteated pPDV089
Method/Materials:
- 3 clones from pPDV089_3F6
- 3 clones from pPDV089_3F8
- 3 clones from pPDV089_2S7
- 3 clones from pPDV089_2S14
- each clone picked 2x: one for growing in LB medium with kanamycin, one in LB medium with ampicillin
- 5 ml LB medium per clone
- storage over night at 37°C and 800 rpm
Further tasks:
- control, whether clones grew in presence of ampicillin but not of kanamycin
Results:
- clones grew in presence of both kanamycin and ampicillin
- kanamycin deletion didn`t work out
79th Labday 2011-08-29
check transformation from yesterday (pSB1C3+mdnABCDE89 and pUP089+mdn-Lib2)
Investigators: Nadine, Steffi
Aim: check transformation
Time: 2011-08-28, 8:20-8:40
Materials:
- Transformations from yesterday (2011-08-28, Kat)
Conclusions:
- transfomations didn´t work
- repeat PCRs
Further tasks:
- repeat PCR for mdnABCDE
- repeat PCR for mdnA-Library
miniprep of several mdn clones
Investigators: Steffi
Time: 2011-08-29, 07:00-09:30
Aim:
- purification of 15x pSB1C3+mdnX plasmids for mdnX biobricks (and screening) from over night cultures (Katharina, 2011-08-28)
Materials
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl Elution buffer
- Check concentration with NanoDrop
- o.n. cultures (from 2011-08-28, Kat):
- pSB1C3+mdnC clone 1
- pSB1C3+mdnC clone 2
- pSB1C3+mdnC clone 3
- pSB1C3+mdnD clone 1
- pSB1C3+mdnD clone 2
- pSB1C3+mdnD clone 3
- pSB1C3+mdnE clone 1
- pSB1C3+mdnE clone 2
- pSB1C3+mdnE clone 3
- pSB1C3+mdnABC clone 1
- pSB1C3+mdnABC clone 2
- pSB1C3+mdnABC clone 3
- pSB1C3+mdnDE clone 1
- pSB1C3+mdnDE clone 2
- pSB1C3+mdnDE clone 3
Results:
- pSB1C3+mdnC clone 1: 434,1 ng/µl
- pSB1C3+mdnC clone 2: 304,4 ng/µl
- pSB1C3+mdnC clone 3: 620,7 ng/µl
- pSB1C3+mdnD clone 1: 282,6 ng/µl
- pSB1C3+mdnD clone 2: 413,4 ng/µl
- pSB1C3+mdnD clone 3: 440,0 ng/µl
- pSB1C3+mdnE clone 1: 543,7 ng/µl
- pSB1C3+mdnE clone 2: 573,5 ng/µl
- pSB1C3+mdnE clone 3: 440,5 ng/µl
- pSB1C3+mdnABC clone 1: 542,3 ng/µl
- pSB1C3+mdnABC clone 2: 267,5 ng/µl
- pSB1C3+mdnABC clone 3: 263,6 ng/µl
- pSB1C3+mdnDE clone 1: 366,5 ng/µl
- pSB1C3+mdnDE clone 2: 440,2 ng/µl
- pSB1C3+mdnDE clone 3: 340 ng/µl
Output:
- plasmids: see results (stored in -20°C: green box, red label: mdn biobricks)
Further Tasks:
- test digest (today)
- sequencing (today?)
test digest of pSB1C3+mdncombinations (several mdn clones/combinations)
Investigators: Steffi
Aim: prove of Insert (mdn combinations)
Materials:
- pSB1C3+mdn-combinations (miniprep from today, see above, Steffi)
- HindIII, PvuII, ClaI, AvaI, HpaI (NEB)
- Buffer 4 and 2 (NEB, 10x)
- BSA
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnC | 3385 | HindIII | 2 | 363, 3022 |
| mdnD | 2944 | PvuII | 4 | 220, 591, 2133 |
| mdnE | 4474 | ClaI | 4 + BSA | 769, 3705 |
| mdnABC | 4852 | HindIII, AvaI | 2 | 363, 836, 3653 |
| mdnDE | 5154 | HpaI | 4 | 1329, 3825 |
Digestion protocol (one enzyme)
- 1 µl DNA
- 0.5 µl enzyme (see table)
- 2 µl 10x buffer
- 16.5 µl pure water
Digestion protocol (two enzymes)
- 1 µl DNA
- 0.5 µl enzyme I
- 0.5 µl enzyme II
- 2 µl 10x buffer
- 16 µl pure water
Digestion protocol (two enzymes and BSA)
- 1 µl DNA
- 0.5 µl enzyme I
- 0.5 µl enzyme II
- 2 µl 10x buffer
- 0.2 µl BSA
- 15.8 µl pure water
- total: 20µl
- 37°C for 1,5 h
Result gel:
| lane | sample | enzymes | volume in µl | Expected size in bp |
| M1 | 1:100 DNA ladder mix, Fermentas | 12 | ||
| 1 | - | - | - | - |
| 2 | mdnC 1 | HindIII | 24 | 363, 3022 |
| 3 | mdnC 2 | HindIII | 24 | 363, 3022 |
| 4 | mdnC 3 | HindIII | 24 | 363, 3022 |
| 5 | mdnD 1 | PvuII | 24 | 220, 591, 2133 |
| 6 | mdnD 2 | PvuII | 24 | 220, 591, 2133 |
| 7 | mdnD 3 | PvuII | 24 | 220, 591, 2133 |
| 8 | mdnE 1 | ClaI | 24 | 769, 3705 |
| 9 | mdnE 2 | ClaI | 24 | 769, 3705 |
| 10 | mdnE 3 | ClaI | 24 | 769, 3705 |
| 11 | mdnDE 1 | HpaI | 24 | 1329, 3825 |
| 12 | - | - | - | |
| 13 | mdnDE 2 | HpaI | 24 | 1329, 3825 |
| 14 | mdnDE 3 | HpaI | 24 | 1329, 3825 |
| 15 | mdnABC 1 | HindIII, AvaI | 24 | 363, 836, 3653 |
| 16 | mdnABC 2 | HindIII, AvaI | 24 | 363, 836, 3653 |
| 17 | mdnABC 3 | HindIII, AvaI | 24 | 363, 836, 3653 |
Further tasks:
- sequencing of mdnC (2+3), mdnD (1+3), mdnE (2)
- repeat PCR of mdnB (wait for new primer), mdnABC and mdnDE
competent cells - E.coli XL1 blue
Investigator: Steffi
Aim: produce competent cells
Materials/Methods:
| TFB I | 1000ml | 200ml |
| 100mM Rubidium Chloride | 12.1 | 2.42g |
| 30mM Potassium Acetate | 2.944 | 0.59g |
| 10mM Calcium Chloride | 1.47 | 0.29g |
| 15% w/v Glycerol (87%) | 150 | 34.5g |
Adjust pH to 5.8 with acetic acid
Filter sterilize the solution
| TFB II | 500ml | 100ml |
| 50mM Rubidium Chloride | 0.6 | 0.121g |
| 10mM MOPS | 1.05 | 0.210g |
| 75mM Calcium Chloride | 5.51 | 1.100g |
| 15% w/v Glycerol (87%) | 75 | 17.24g |
Adjust pH to 7.0 with KOH
Filter sterilize the solution
Work always sterile and cold and speedy!
- All volumes deal with the common cellline!
- Prepare 80 Eppis (1,5µl)
- get liquid nitrogen
- prepare 5 ml LB-Medium with the specific antibiotic (for XL1-blue: Tet), inoculate and incubate over night
- prepare 200 ml LB-Medium with the specific antibiotic, inoculate with 2 ml of the over-night-culture
- grow while shaking at 37°C, 190 rpm to an OD600 at 0,4-0,6
- keep cell suspension in sterile falcons (50 ml) 10 min on ice, then centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend on ice with 40 ml icecold TFB I and keep 10 min on ice
- centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend pellet in 8 ml TFB II
- aliquot in Eppis: 50µl per tube and store immediately at liquid nitrogen and afterwards at -80 °C
Results:
- 80 tubes XL1 blue for transformation(50 µl competent cells at -80°C)
Further tasks:
check by transformation and check the resistance on agar plates with different antibiotics
check resistance of competent cells - E.coli XL1 blue
Investigator: Katharina, Steffi
Aim: check resistance of competent XL1 blue-cells on agar plates with different antibiotics
Materials/Methods:
- competent XL1 blue-cells from 2011-08-27 and 2011-08-29 (Steffi)
- LB-plates with Tetracycline, Chloramphenicol, Kanamycine, Ampicillin
- plating 50 µl on agar plates
- incubate at 37°C over night
Further tasks:
control agar plates
Oligo-Fillin for mdnA-Library (repetition)
Time: 2011-08-29, 8:30-11:30
Investigators: Nadine
Materials
- Primer, 25 µM :
- # 76
- # 74: o_foc_library_2
- # 72: o_loop_con_switch
- Klenow-Buffer 10X
- Klenow Fragment
- dNTPs
- water
- Agarose (BioRad)
- 1x TAE
- gel red
- loading dye (promega)
- 100bp DNA ladder (Fermentas)
- 1kb DNA ladder (Promega)
Protocol:
- Reaction mix
- 1 µl fw-Oligonucleotide (#76)
- 1 µl rev-Oligonucleotide (#74 or #72)
- 2 µl dNTPs (10 mM)
- 2 µl Klenow-Fragment
- 14 µl water
- total volume: 20 µl
2. PCR program
- name: Fillin
- 3 min 94 °C
- 0.3°C per s (94°C-37°C)
- addition of 0.5 µl Klenow Fragment
- press enter
- 1hr 37°C
Agarose gel:
1.5 %: 0.75 g in 50 ml TAE
Results:
- Output:
- mdnA-Lib2, Nad, 29.8.11 (~170bp)
- mdnA-loop, Nad, 29.8.11 (~170bp)
Further tasks:
- PCR purification
sequencing of TEV and 14_3C clones
Aim: get sequences
Methode:
send samples to GATC
Further Tasks:
Restriction of pUP089 (vector) and Lib-2/loop(Insert/PCR)
Investigator: Niels
Aim: generate vector and insert for ligation and transformation to generate a Libary
Materials/Methods:
- pub089 (A)
- 5 µl DNA
- 0,5 µl Ehe I
- 0,5 µl Aat II
- 3 µl 10x Buffer
- 21 µl water
- total volume: 30µl
- pub089 (B)
- 1 µl DNA
- 0,5 µl Ehe I
- 0,5 µl Aat II
- 3 µl 10x Buffer
- 25 µl water
- total volume: 30µl
- Lib-2 (A)
- 20 µl DNA
- 0,5 µl Aat II
- 3 µl 10x Buffer
- 6,5 µl water
- total volume: 30µl
- Lib-2 (B)
- 1 µl DNA
- 0,5 µl Aat II
- 3 µl 10x Buffer
- 25,5 µl water
- total volume: 30µl
- Loop
- 1 µl DNA
- 0,5 µl Aat II
- 3 µl 10x Buffer
- 25,5 µl water
- total volume: 30µl
over night
Further tasks:
- purification
- dephosphorylation
- ligation
- transformation
PCR of complete mdn cluster for biobrick-construction
Investigators: Niels, Nadine
Material:
- pARW089
- pARW071
- dNTPs
- primer 77, 78, 79, 80
- HF Phusion Buffer 5x* Phusion Polymerase
Method:
- 2µl pARW089, 1µl pARW071, respectively
- 1µl dNTPs
- 2µl forward primer (10mM)
- 2µl reverse primer (10mM)
- 10µl HF Phusion Buffer 5x
- 0.5 µl Phusion Polymerase
- ad 50 µl water
- reaction
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 95°C | 30 s |
| Annealing | 59°C | 60 s |
| Extension | 72°C | 3,5 min |
| DenaturationII | 95°C | 30 s |
| AnnealingII | 59°C | 60 s |
| ExtensionII | 72°C | 3.5 min |
| Final extension | 68°C | 5 min |
Result:
- three different PCR samples
- PCR sample 1: pARW071 + primer 77/78 = mdnABCDE71
- PCR sample 2: pARW089 + primer 77/79 = mdnABCDE89+T7
- PCR sample 3: pARW089 + primer 77/80 = mdnABCDE89
Further Tasks:
- gel electrophoresis
- PCR clean up
- restriction enzyme digestion
- ligation
- transformation
Alignment of sequenced pPDV089 and model of pPDV089
Investigator: Sabine
Aim: control of generated phage display vector pPDV089
Materials/Methods:
- file sent from GATC
- software Genious
Results:
- 4 positive clones, pPDV089 containing mdnA and geneIII (3F6, 3F8, 2S7, 2S14)
Further task:
- deletion of kanamycin gene
PCR of geneIII and mdnA
Investigators: Sabine
Aim:
- amplification of mdnA and geneIII for cloning into pSB1C3
- amplification of geneIII for cloning into pARW089
Reaction Components:
- 2 µl/ 20 ng geneIII (cut out from pARW089
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl primer pf_geneIII_EheI_XbaI_NgoMIV
- 1 µl primer pr_geneIII_iGEM_AatII
- 5 µl 10x PCR Buffer S
- 39,75 µl water
- 5 µl/ 50 ng pak bla KDIR (mdnA)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl primer pf_mdnA_iGEM_EheI
- 1 µl primer pr_mdnA_iGEM_AatII
- 5 µl 10x PCR Buffer S
- 36,75 µl water
Further tasks:
- analytic gel electrophoresis
- purification
- digestion
80th Labday 2011-08-30
Agarose gel of PCR of complete mdn cluster for biobrick-construction
Time: 2011-08-27, 08:00-09:00
Investigators: Nadine, Steffi
Aim:
- prove insert of pARW071/ pARW089 with complete mdn-cluster (PCR-product from 2011-08-29, Nadine)
Materials:
- Agarose (BioRad)
- 1x TAE
- gel red
- loading dye (promega)
- 1kb DNA ladder (Promega)
Protocol:
- 1 x 1%: 0.5 g agarose + 50 ml 1xTAE buffer, 2 µl gel red
Results:
- gel
| lane | sample | volume in µl | Expected size in bp |
| M1 | 1kb DNA ladder (Promega) | 10 | - |
| 1 | mdnABCDE71 | 2 | |
| 2 | mdnABCDE89 | 2 | |
| 3 | mdnABCDE89+T7 | 2 |
Conclusions:
- PCR didn`t work
Further Tasks:
- repeat PCR
- gel electrophoresis
- PCR clean up
- restriction enzyme digestion
- ligation
- transformation
Repeat PCR of complete mdn cluster for biobrick-construction
Investigators: Nadine, Vanessa, Steffi
Aim: repeat PCR of complete mdn cluster for biobrick-construction from 2011-08-29
Material:
- pARW089
- pARW071
- dNTPs
- primer 77, 78, 79, 80
- Bio-X-ACT Long DNA Polymerase (Bioline)
Method:
- 1µl pARW089 (~ 3.3ng) , 1µl pARW071 (~ 5ng)
- 0.5µl dNTPs (100 mM)
- MgCl2 (50 mM)
- 1.5µl forward primer (100µM)
- 1.5µl reverse primer (100µM)
- 5µl OptiBuffer 10x
- 1µl Bio-X-ACT Long DNA Polymerase (Bioline)
- add 50 µl water
- reaction
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 95°C | 30 s |
| Annealing | 47°C | 60 s |
| Extension | 72°C | 6,5 min |
| DenaturationII | 95°C | 30 s |
| AnnealingII | 68°C | 60 s |
| ExtensionII | 72°C | 6.5 min |
| Final extension | 68°C | 5 min |
- first steps: 15x
- second step: 25x
- program: igbBio2
Result:
- three different PCR samples
- PCR sample 1: pARW071 + primer 77/78 = mdnABCDE71
- PCR sample 2: pARW089 + primer 77/79 = mdnABCDE89+T7
- PCR sample 3: pARW089 + primer 77/80 = mdnABCDE89
Further Tasks:
- gel electrophoresis
- PCR clean up
- restriction enzyme digestion
- ligation
- transformation
Gel extraction and purification of pUP089 (vector) and Lib-2/loop(Insert/PCR)
Investigators: Steffi
Materials:
- Restriction products of pUP089 (vector, 10196 bp) and Lib-2/loop(Insert/PCR) from Niels (2011-08-29)
- Agarose (BioRad)
- 1x TAE
- gel red
- loading dye (promega)
- 1kb DNA ladder (Promega)
- 100bp DNA ladder (Fermentas)
Protocol:
- 1 x 1%: 0.5 g agarose + 50 ml 1xTAE buffer, 2 µl gel red
- 1 x 1.5 %: 0.75 g in 50 ml 1xTAE, 2 µl gel red
Results:
- gel 1 (1.5%)
| lane | sample | volume in µl | Expected size in bp |
| M1 | 1kb DNA ladder (Fermentas) | 10 | - |
| M2 | 100bp DNA ladder (Fermentas) | 10 | - |
| 1 | Lib-2 (A) | 30 | 161 |
| 2 | Lib-2 (B) | 30 | 161 |
| 3 | Loop | 30 | 161 |
- gel 2 (1%)
| lane | sample | volume in µl | Expected size in bp |
| M1 | 1kb DNA ladder (Fermentas) | 10 | - |
| 1 | pUP089 (A) | 30 | 10035 |
| 2 | pUP089 (B) | 30 | 10035 |
Conclusions:
- cut out bands from Lib-2 (B), Loop, pub089 (A) and pub089 (B)
Gel extraction
- Macherey Nagel - NucleoSpin Extract II, protocol for DNA extraction from agarose gels
- Elution with 50µl NE-Buffer
NanoDrop: Concentrations
- Lib-2 (B) - digested - purified: 5,8 ng/µl
- Loop - digested - purified: 3,9 ng/µl
- pUP089 (A): 12,9 ng/µl
- pUP089 (B): 23,4 ng/µl
Further tasks:
- dephosphorylation of pub089 (B)
- ligation
- transformation
Dephosphorylation and Ligation of pUP089 (vector) and Lib-2/loop(Insert/PCR)
Investigators: Nadine, Steffi
Dephosphorylation of pUP089 (B)
- add 5µl Multicore 10x Buffer (Promega)
- add 1µl Thermosensitive Alkaline Phosphatase (Promega)
- incubate at 37°C for 15 min
- heat inactivation at 74°C for 15 min
Ligation
- purified and dephosphorylated pUP089 (B) from 2011-08-30 (vector)
- purified Lib-2 (B) (insert)
- purified Loop (insert)
| Insert | [µl] | Vector | [µl] |
| Lib-2 (B) | 1.8 | pUP089 | 10 |
| Loop | 2.7 | pUP089 | 10 |
- 3 µl 10x Buffer
- 1 µl T4 Ligase
- add 30 µl H2O
- 1h, RT
Further tasks:
- transformation
Transformation of pUP089+library2
Investigators: Niels
Aim: Transformation of Ligation
Materials:
- competent E. coli cells (XL1-Blue, 2011-08-27, 1xSteffi, 2xSabine)
- ligation products: pUP089+Lib2 (2011-08-30, Steffi)
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 90 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Kan)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Output:
- 3 plates w/ Kan: mdn-lib2
sequencing of mdnC, mdnD and mdnE BioBrick
Time: 2011-08-30,14:00-14:30
Investigators: Nadine
Materials
Sequencing by GATC
- C2 (mdnC Klon 2, Steffi, 2011-8-29)
- C3 (mdnC Klon 3, Steffi, 2011-8-29)
- D1 (mdnD Klon 1, Steffi, 2011-8-29)
- D3 (mdnD Klon 3, Steffi, 2011-8-29)
- E2 (mdnE Klon 2, Steffi, 2011-8-29)
check plates - resistance of competent E.coli XL1 blue cells
Investigators: Nadine, Steffi
Aim: check resistance of competent cells - E.coli XL1 blue (from 2011-08-27 and 2011-08-29, Steffi)
Materials:
- agar plates from competent cells - E.coli XL1 blue from 2011-08-29 (Kat/ Ste)
Results:
- all competent cells - E.coli XL1 blue grow on agar plates with Tetracycline
- no E.coli XL1 clones on agar plates with Ampicillin, Kanamycine, Chloramphenicol
Conclusions:
- competent cells - E.coli XL1 blue work and can be used for transformation
Assembly PCR for TEV and 14_3C
Aim: get the side-directed mutated TEV and 14_3C fragment
Methode:
Primer TEV:
(1) f_TEV_iGEM
(2) r_TEV_iGEM_BamHI
Primer 14_3C:
(1) f_14_3C_iGEM
(2) r_14_3C_iGEM_BamHI
Methode:
PCR
- Template: 1 µL (TEV or 14_3C <10ng)
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 5 µL 10 x Amplification buffer S
- 2 µL 25 mM MgCl2
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer)
- 35,5 µL of pure water
- 0,5 µL TaqPol
Program:
- Denat: 4 min 94°C
- 5x:
Denat: 1 min 94°C
Anneal: 1 min 52°C
Extend: 1 min 72°C
- 25x:
Denat: 1 min 94°C
Anneal: 1 min 69°C
Extend: 1 min 72°C
- Final Extend: 10min 72°C
Further Tasks:
PCR purification to check the correct sizes of fragments
Digestion of created pPDV089 for deletion of kanamycin gene
Investigators: Sabine
Aim:
- inactivation of kanamycin resistence gene
- reason: helper plasmid contains also kanamycin restistence gene
- enable selection of E. coli cells containing both helper plasmid and phage display vector
Reaction components:
- 4 µl/2µg pPDV089 (from clones 3F6, 3F8, 2S7 and 2S14)
- 1 µl restriction enzyme NsiI
- 2 µl buffer 3
- 13 µl water
Reaction conditions:
- 3 h at 37°C
Digestion of PCR products geneIII and mdnA for cloning into pSB1C3
Investigator: Sabine
Aim: cloning of mdnA, geneIII and mdnA/geneIII into pSB1C3
Material/Method:
- 15 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 2 µl NEB 10x buffer 2
- 0,2 µl BSA
- 0,8 µl water
- 1,5 h, 37°C
- 15 µl PCR product geneIII
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 2 µl NEB 10x buffer 2
- 0,8 µl water
- 0,2 µl BSA
- 1,5 h, 37°C
- 15 µl PCR product geneIII
- 1 µl restriction enzyme NgoMIV
- 1 µl restriction enzyme PstI
- 3 µl NEB 10x buffer 1
- 1,5 h, 37°C
- 15 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme AgeI
- 3 µl NEB 10x buffer 1
- 0,2 µl BSA
- 1,5 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation of mdnA and geneIII into pSB1C3
Digestion of PCR product geneIII and vector pARW089
Investigator: Sabine
Aim: pARW089 containing geneIII but not mdnA
Material/Method:
- 15 µl PCR product geneIII
- 1 µl restriction enzyme SfoI
- 1 µl restriction enzyme AatII
- 2 µl NEB 10x buffer 4
- 1 µl water
- 1,5 h, 37°C
- 6 µl/2µg pARW089
- 1 µl restriction enzyme SfoI
- 1 µl restriction enzyme AatII
- 1 µl NEB 10x buffer 4
- 1 µl water
- 3 h, 37°C
Further Task:
- purification
- ligation
Ligation of geneIII into pARW089
Investigator: Sabine
Aim: cloning of geneIII into pARW089
Material/Method:
- 1,5 µl PCR product geneIII (14,5 µg/µl)
- 15,5 µl pARW089 (11 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Re-ligation of NsiI-digested pPDVs089
Investigator: Sabine
Aim: create pPDV089 without kanamycin resistence
Material/Method:
- 17 µl NsiI-digested pPDVs089 (3F6, 3F8, 2S7 and 2S14, DNA from upper and lower bands)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Ligation of geneIII and mdnA into pSB1C3
Investigator: Sabine
Aim: pSB1C3 containing mdnA, geneIII and mdnA/geneIII
Material/Method:
- 12,5 µl PCR product geneIII (10 µg/µl)
- 4,5 µl pSB1C3 (41 ng/µl)
- 2 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 1 h at room temperature
- 10,8 µl PCR product mdnA (7 µg/µl)
- 6,2 µl pSB1C3 (41 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
- 4,5 µl PCR product mdnA (7 µg/µl)
- 10 µl PCR product geneIII (7 µg/µl)
- 2,5 µl pSB1C3 (41 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Transformation of created vectors in E. coli
Investigator: Sabine
Aim:amplification of vectors
Material:
- pPDV089 without kanamycin resistence (3F6, 3F8, 2S7 and 2S14)
- pARW089 containing only geneIII (no mdnA)
- pSB1C3 containing mdnA, geneIII or mdnA/geneIII
- agar plates containing chloramphenicol (pSB1C3)
- agar plates containing kanamycin, ampicillin(pPDV)
- agar plates containing kanamycin (pARW089)
Method:
- addition of 4 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
control cell clones
gel electrophoresis of TEV and 14_3C
Aim: check sizes
Method:
80 V
Further Tasks:
PCR purification
81th Labday 2011-08-31
Ligation of geneIII into pARW089
Investigator: Sandrina, Laura
Aim: cloning of geneIII into pARW089
Material/Method:
- 2 µl PCR product geneIII (14,5 ng/µl)
- 15 µl pARW089 (16 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Transformation of pARW089 containing only geneIII (no mdnA) in E.coli
Investigator: Sandrina, Laura
Aim:amplification of vectors
Method:
- addition of 4 µl ligation reaction to XL1-blue cells
- incubation 20 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml kanamycin
- storage over night at 37°C
Further tasks:
control cell clones
overnight culture of picked E. coli clones transformed with pPDV089 to control deletion of kanamycin gene, pARW089 containing only geneIII (no mdnA), pSB1C3 containing mdnA, geneIII or mdnA/geneIII
Investigators: Sandrina, Laura
Aim: control deletion of kanamycin gene in created pPDV089, control ligation of geneIII into PARW089, control ligation of mdnA, geneIII and mdnA/geneIII into pSB1C3
Method/Materials:
- 5 clones from pPDV089_2S14 (ampicillin)
- 5 clones from pARW089 with geneIII (kanamycin)
- 5 clones from pSB1C3 with mdnA (chloramphenicol)
- 5 clones from pSB1C3 with geneIII (chloramphenicol)
- 5 clones from pSB1C3 with mdnA/geneIII (chloramphenicol)
- 5 ml LB medium per clone
- storage over night at 37°C and 800 rpm
Further tasks:
- test digestion
over night cultures of pSB1C3+mdnABC/mdnDE
time: 2011-8-31, 18:00
Investigators: Nadine
Aim: check colonies for correct plasmid
Materials:
- agar plates from 2011-8-25, Niels (fridge):
- pSB1C3+mdnABC
- pSB1C3+mdnDE
- LB medium
- chloramphenicol (25 mg/ml)
Protocol:
- 10 x: 10 ml LB medium + 10 µl chloramphenicol
- pick colony from plate (from each plate 5 colonies, respectively)
- transfer to LB medium
- incubate over night in 37°C shaker (200 rpm)
Output:
- over night cultures:
- pSB1C3+mdnABC
- clone 3
- clone 4
- clone 5
- clone 6
- clone 7
- pSB1C3+mdnDE
- clone 1
- clone 2
- clone 3
- clone 4
- clone 5
Further tasks:
- miniprep
- test digest (for protocol see 2011-8-29)
Assembly PCR for 14_3C, PCR of AraC and TEV
Aim: get the side-directed mutated TEV and 14_3C fragment
Method:
Primer TEV:
(1) f_TEV_iGEM
(2) r_TEV_iGEM_BamHI
Primer 14_3C:
(1) f_14_3C_iGEM
(2) r_14_3C_iGEM_BamHI
Methode:
PCR
- Template: 1 µL (TEV or 14_3C <10ng)
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 5 µL 10 x Amplification buffer S
- 2 µL 25 mM MgCl2
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer)
- 35,5 µL of pure water
- 0,5 µL TaqPol
Program:
- Denat: 4 min 94°C
- 5x:
Denat: 1 min 94°C
Anneal: 1 min 52°C
Extend: 1 min 72°C
- 25x:
Denat: 1 min 94°C
Anneal: 1 min 69°C
Extend: 1 min 72°C
- Final Extend: 10min 72°C
Further Tasks:
PCR purification to check the correct sizes of fragments
 "
"