Team:Peking S/project/nonb
From 2011.igem.org
Template:Https://2011.igem.org/Team:Peking S/bannerhidden Template:Https://2011.igem.org/Team:Peking S/back2
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Non-Boolean Population
Introduction|Design|Comparator
Introduction
By compartmentalizing molecular components of a complex genetic circuit or a pathway, engineered microbial consortia may perform functions that are difficult for monocultures via differentiation and communication. Also, due to metabolic differentiation and possibly an appropriate community structure (e.g., communities without strict competitive hierarchies), microbial consortia may show better endurance in changeable environment.
Recent efforts to engineer microbial consortia primarily focused on either multi-step function performing or population interaction centered dynamics. Apart from several leading attempts in building basic Boolean elements, non-Boolean population dynamics appear to be versatile in both trends. Bidirectional communication-based population interactions (e.g. the synthetic prey-predator ecosystem) have been achieved, but a tri-population circuit with double-bidirectional communication would be an adventurous attempt. It is also an appropriate demonstration for our ‘chemical wire’ toolbox.
Inspired by the fact that integration and comparison of signals is less frequently involved in the construction of intercellular genetic networks, we also hope to fill this blank with our small-RNA based comparator in the monitoring cells.
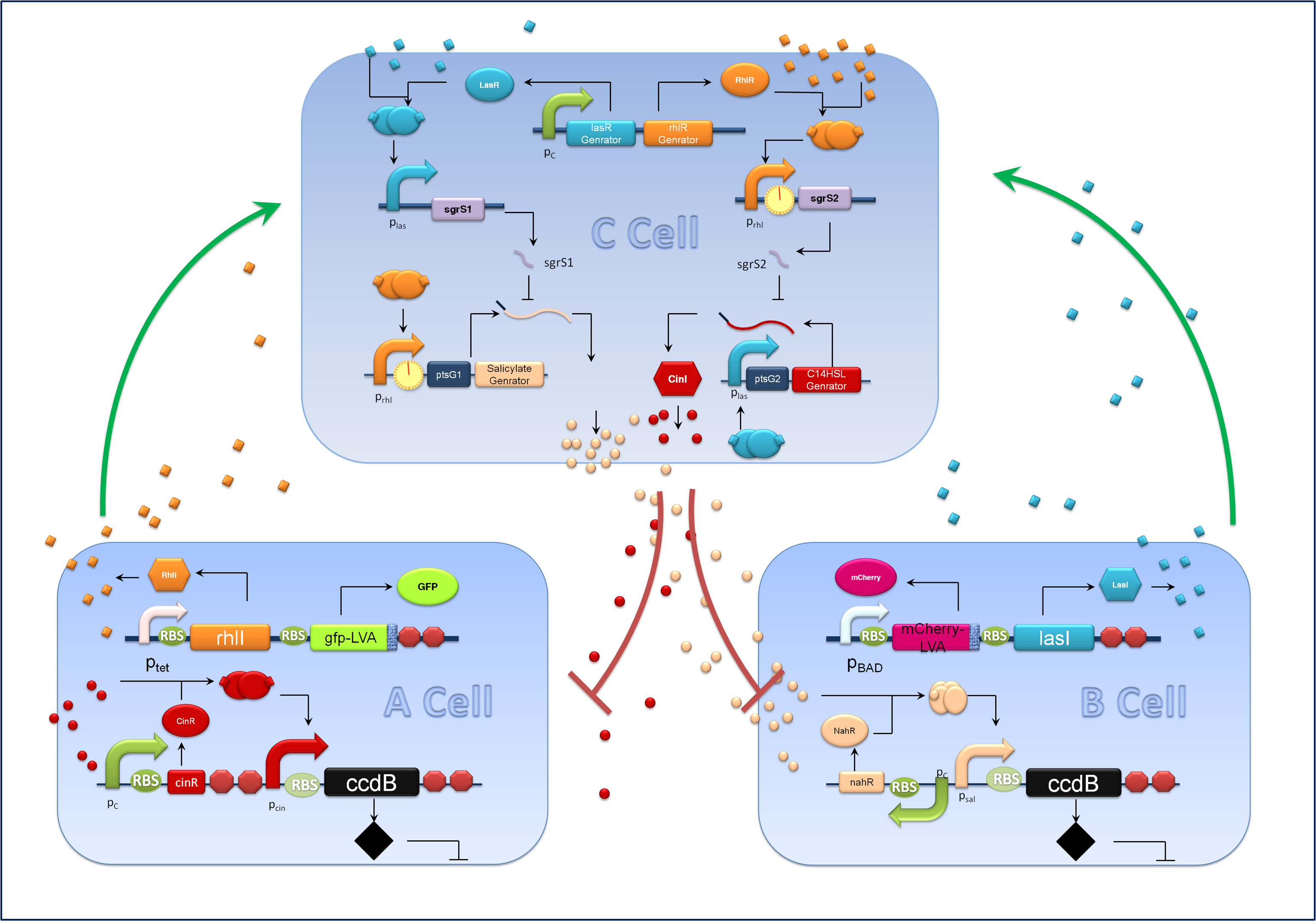
Figure 1. Schematic of our population density balancer.
Aiming at a hierarchal regulation of population growth, signaling molecules report cell densities of the two competing populations and the corresponding feedback signals are essential. Here we harnessed two quorum sensing modules, RhlI/RhlR from Pseudomonas aeruginosa and CinI/CinR from Rhizobium leguminosarum to enable two-way communications between A cells and C cells. Meanwhile the rewired salicylate regulated PchBA/NahR module and the LasI/LasR quorum sensing devices from Pseudomonas aeruginosa are utilized for B-C bidirectional communication. CinI/CinR & PchBA/NahR systems are carefully characterized in our ‘chemical wire’ toolbox module and all four signaling systems are validated for their orthogonality. Check our ‘chemical wire’ toolbox page for more information.
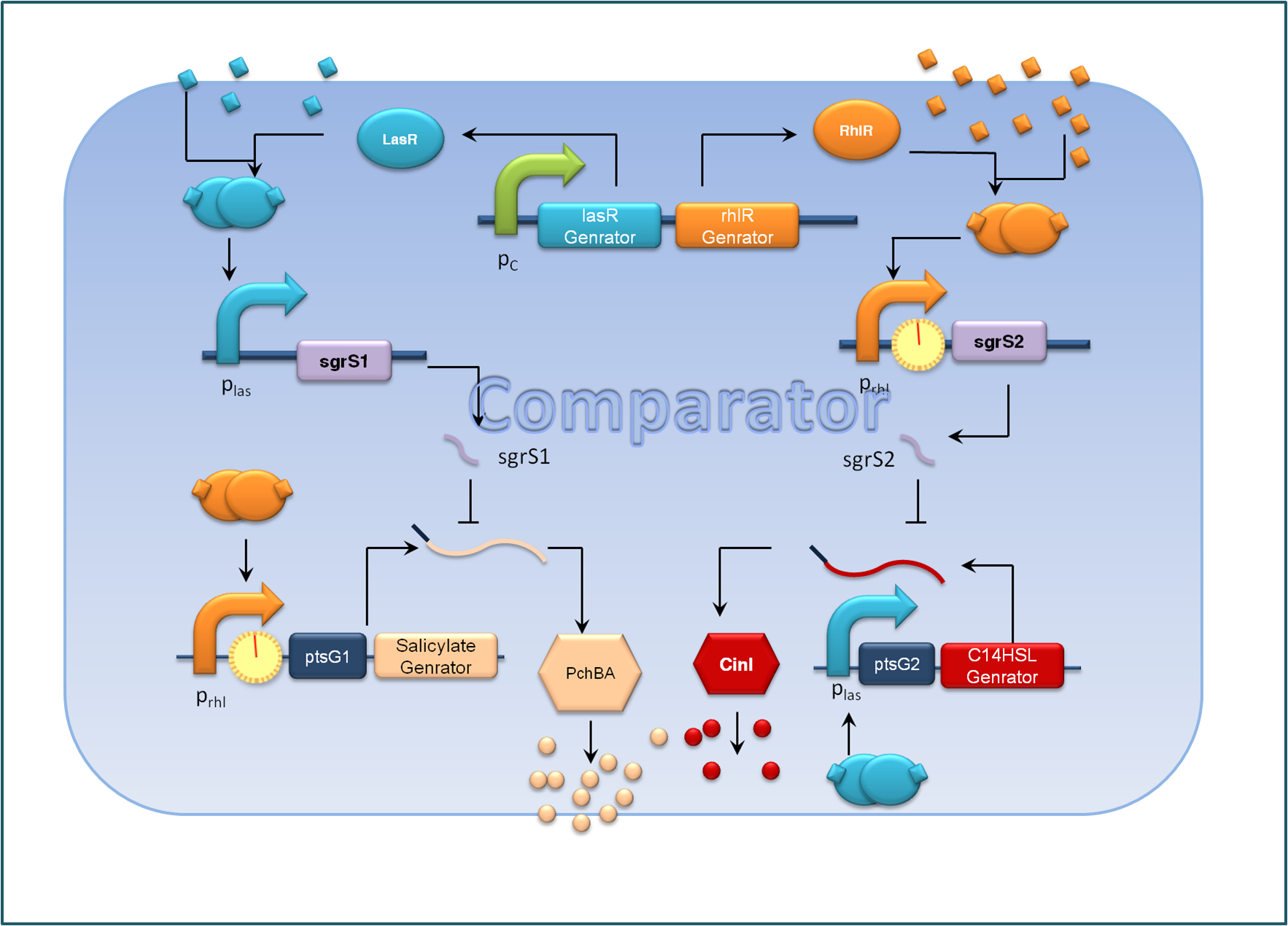
Figure 2. Expected Function of the comparator, C cells
For the sake of effective signal integration, we designed a small RNA based comparator, which is composed of two engineered orthogonal small RNA-mRNA 5’ untranslated region(UTR) pairs. With small RNA from one pair and the target mRNA from the other pair under the control of same inducible promoters, the two pair of RNA regulators interact with each other antagonistically. Under the conditions of A cells dominating, excess C4HSL molecules synthesized by RhlI would lead to a relative overexpression of sgrS2 small RNA regulators, resulting in the silence of ptsG2-cinI mRNA and the consequent lower concentration of C14HSL. At the meantime, the expression of sgrS1 and ptsg1-pchBA remain the same and the overall result is a higher output concentration of salicylate. This seems to be a by-pass signaling molecule transition, yet the comparison of relative signal intensity is hard to achieve otherwise`.
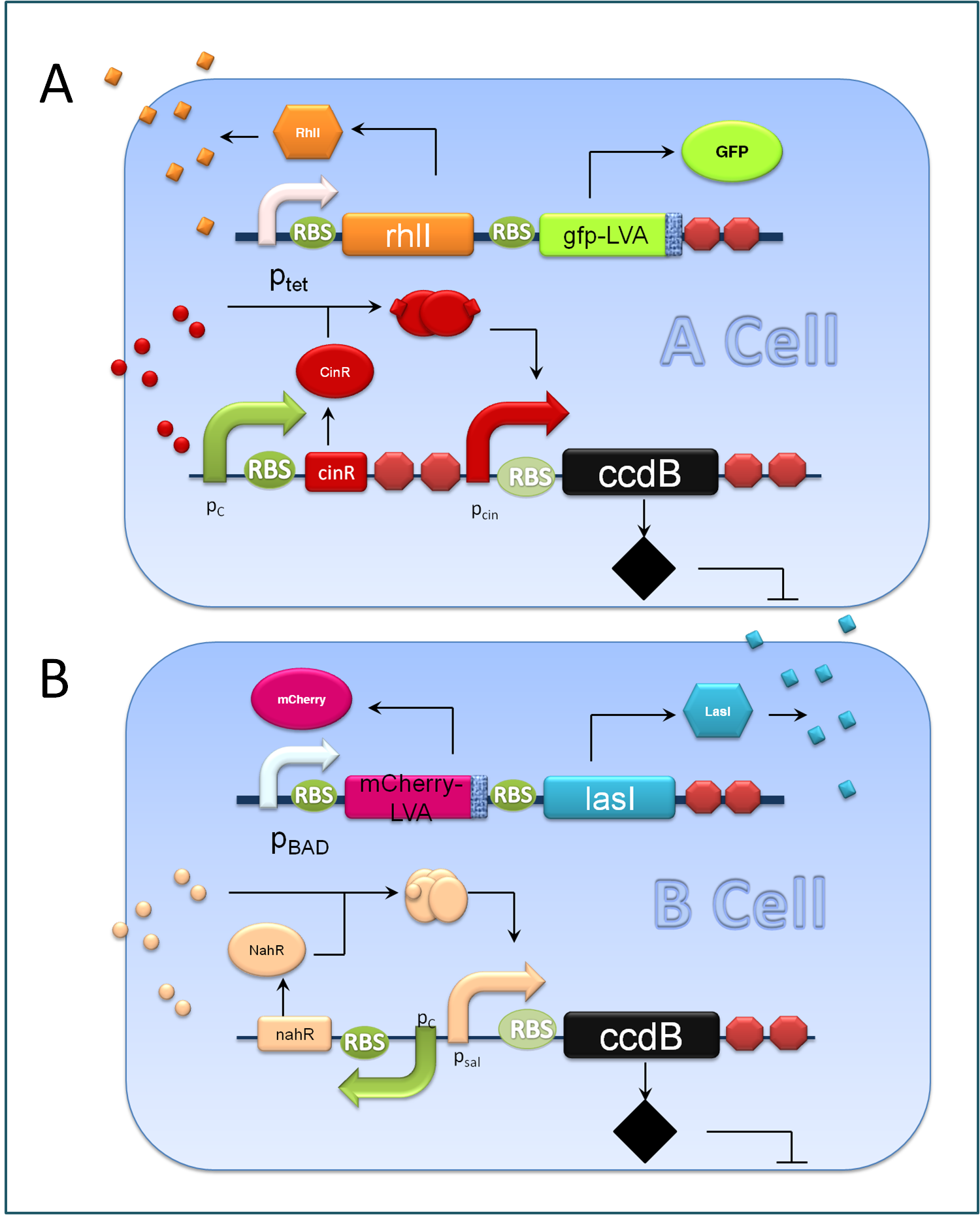
Figure 3. Components of A&B Cells
A cells and B cells basically consist of two discrete modules: a reporting module and an inducible suicide device. The reporting module constitutively express distinct signaling molecules as well as fluorescent proteins and consequently the total concentration of the molecule in culture broth together with the fluorescence strength will clearly indicate the corresponding population density. Inducible promoters in this module are specifically designed for adjusting. The suicide module is directly regulated by chemical signals from C cells, as Figure 3 illustrates.
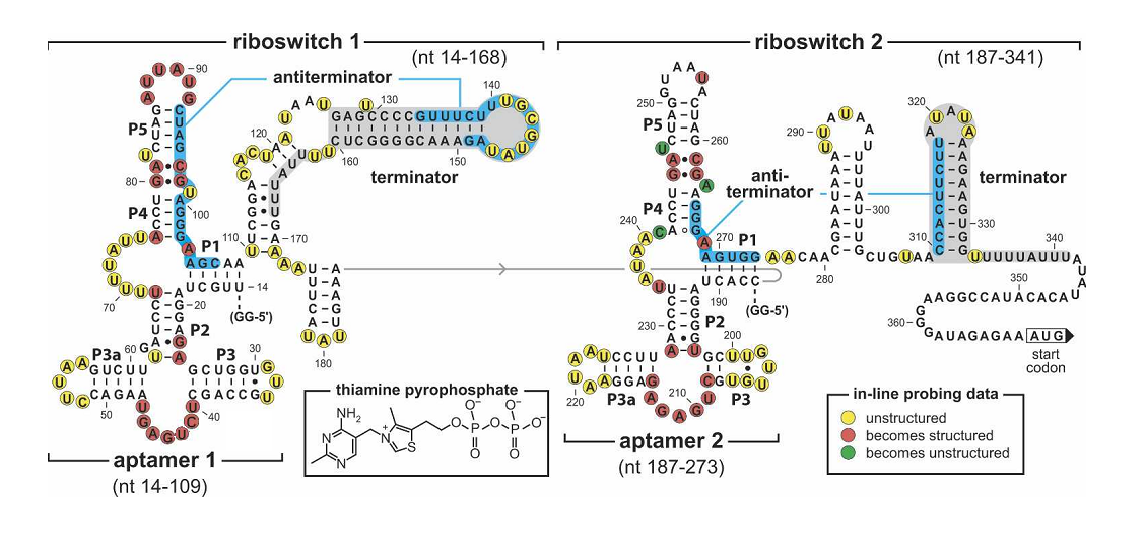
To make our balancer tunable, we introduced a tandem riboswitch responding to different thiamine pyrophosphate (TPP) concentrations. A sufficient TPP concentration results in transcription pre-termination, and thus downstream gene expression is inhibited to some extent. The transcription regulator is placed right after the rhl activated promoters in our system, as shown in Figure 2.
Figure 4. Sequence and the secondary structure of the tandem TPP riboswitch (Rüdiger Welz and Ronald R. Breaker,2007). The riboswitch carry mutations in the junction in the first aptamer(G41C, A42U) disrupting TPP binding.
An ordinary differential equation (ODE) model was built to simulate population behaviors of our system. The results predicted that our population dynamic balancer can control A(B) cell with arbitrary proportion and the whole system always gets to a steady state without oscillation. Click to check our model page for more information.
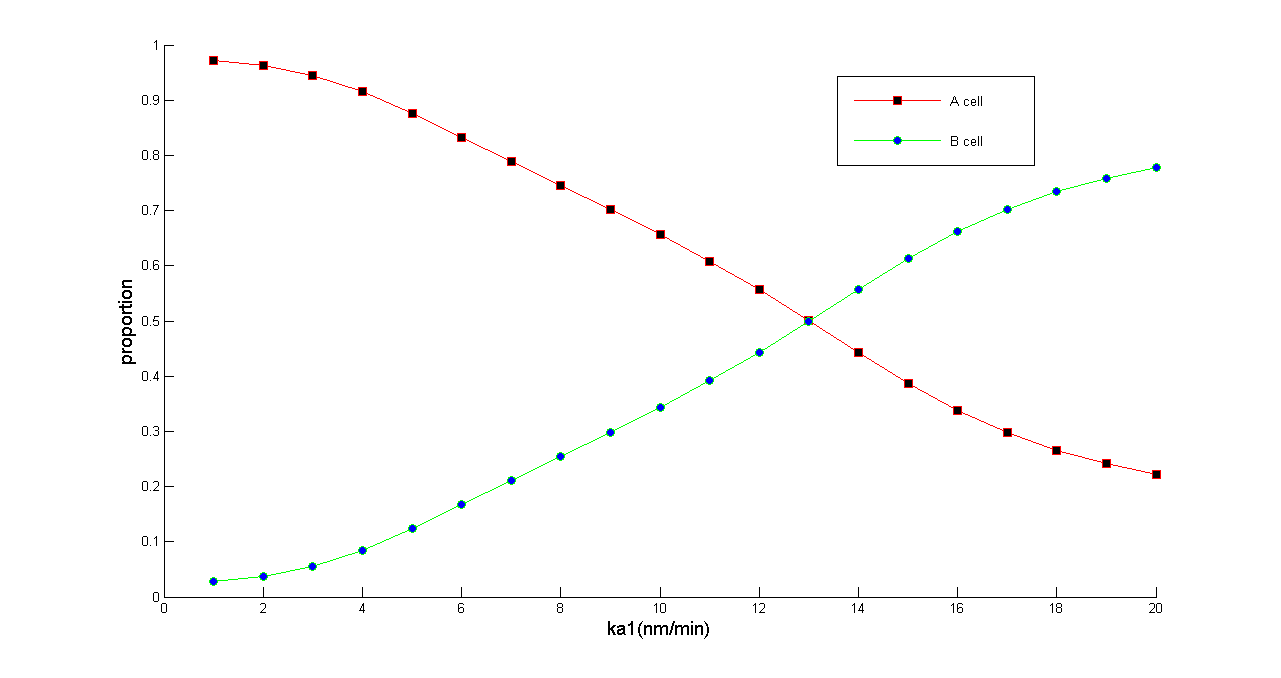
Figure 5 Normalized dose-response curve from the ODE simulation. Ka1 stands for transcription rate of ptsG1 and Salicylate Generator. (in bench work, addition of thiamine pyrophosphate, TPP, can modulate this parameter) As Ka1 vary from 1-20 nmol•min^-1,the ratio of A & B cell populations may be arbitrarily modulated from 0.2 to 1
In terms of engineering communication with dedicated molecules, population behavior is much density-dependent unless the system spontaneously reaches homeostasis and the ability to maintain consortia with unsteady structure is challenged. Taken all these considerations, we designed a general microfluidic platform for cell-cell communication in chemostat. Our A&B cell co-culture showed tunable population ratio dynamics in our microfluidic device. Check the Microfluidic device page for more information.
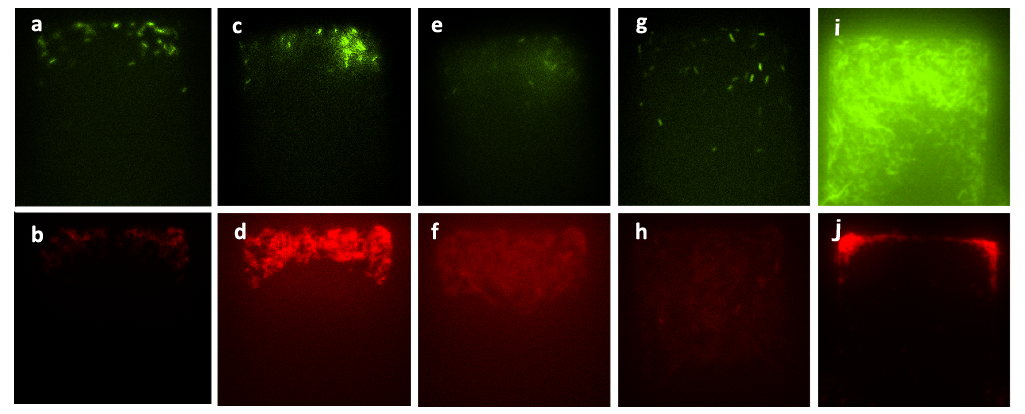
Figure 6. Figure (a) to Figure (h) show the growth of the mixed culture (A cells together with lasI-deficient B cells, referred to as ΔB cells) with inducer generated by CinI (with the initial concentration) inoculated in the upper layer (with the amplification factor of 20, the size of chambers 120μm×120μm, and the intruding velocity of 10μL/h). Photos were taken every one hour, and the time interval lasted three hours. A. cells, which were indicated by GFP they expressed, did not grow during this time interval, while B. cells grew vigorously till they filled up the chamber. Figure (a) to Figure (h) illustrate the fact that the inducer generated by CinI could only repress the growth of A. cells. Figure (i) and Figure (j) demonstrate the final state of this mixed culture’s growth with salicylate (with the concentration of 10^(-3) mol/L )inoculated in the upper layer, which acted as the inducer designed for inducing the expression of CcdB in ΔB cells( also with the amplification factor of 20, the size of chambers 120μm×120μm, and the intruding velocity of 10μL/h). cells. In this case, A. cells finally filled up the chamber, while B. cells kept small population number after three hours’ cultivation.
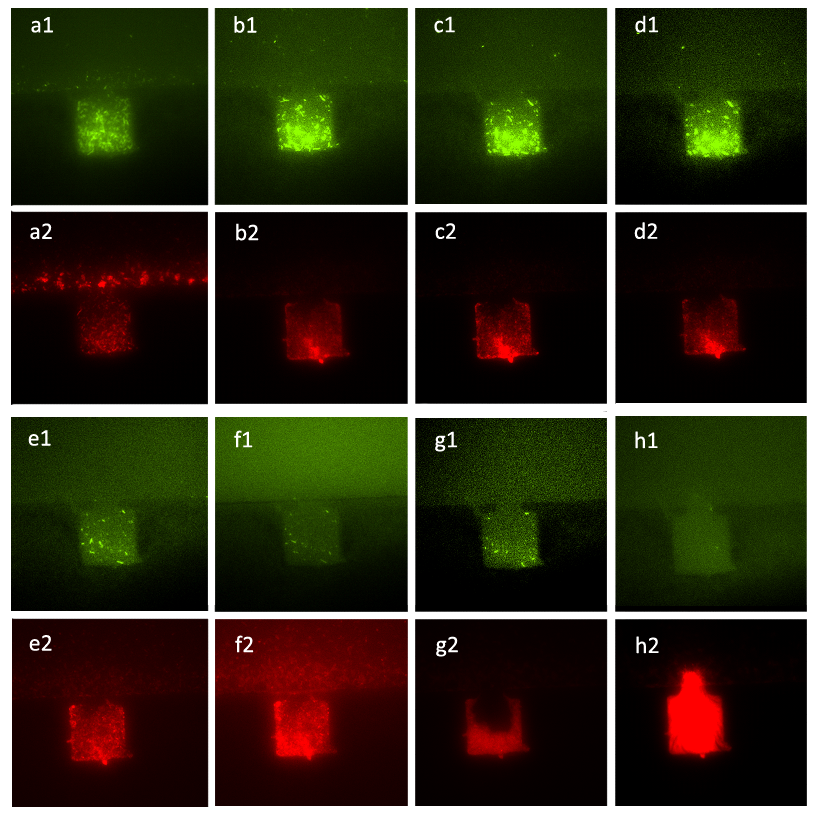
Figure 7. Figure (a_1) and (a_2) show the initial state of the mixed culture in the chamber, the number of A cells and that of ΔB cells were almost the same. Then we inoculated the mixture of salicylate with the concentration of 10^(-3) mol/L and the solution which was diluted in fold of 10^5 from the initial solution of inducer generated by CinI, the inducers for A cells and ΔB cells. Figure (b_1), (b_2), (c_1), (c_2), (d_1) and (d_2) show that the ratio of population number of A. cells to that of B. cells had changed from the initial state and stayed the same during the two hours, which means that the culture came to a steady state under the control of inducers. Figure (e_1), (e_2), (f_1), (f_2), (g_1), (g_2), (h_1) and (h_2) illustrate the growth of the mixed culture with the mixture of salicylate with the concentration of 10^(-4) mol/L and the solution which was diluted with nutrient solution in fold of 10^4 from the initial solution of inducer generated by CinI. During this time interval, ΔB cells grew fast while A cells were extruded from chamber. In Figure (g_1) and (g_2), the number of ΔB cells decreased a little, mainly because the time-delayed effect of the control of salicylate, but the population soon recovered from the effect. Finally, in Figure (h_1) and (h_2), ΔB cells nearly filled up the whole chamber while there were few A. cells in the same chamber.
To sum up, we featured our system with such specifications:
Multiple ‘chemical wires’ utilization
Multi-population interactions and signal integration
Tunable population behavior
Theoretical and physical device supporting
 "
"