Team:Paris Liliane Bettencourt/Notebook/2011/07/23/
From 2011.igem.org
Camille & Danyel
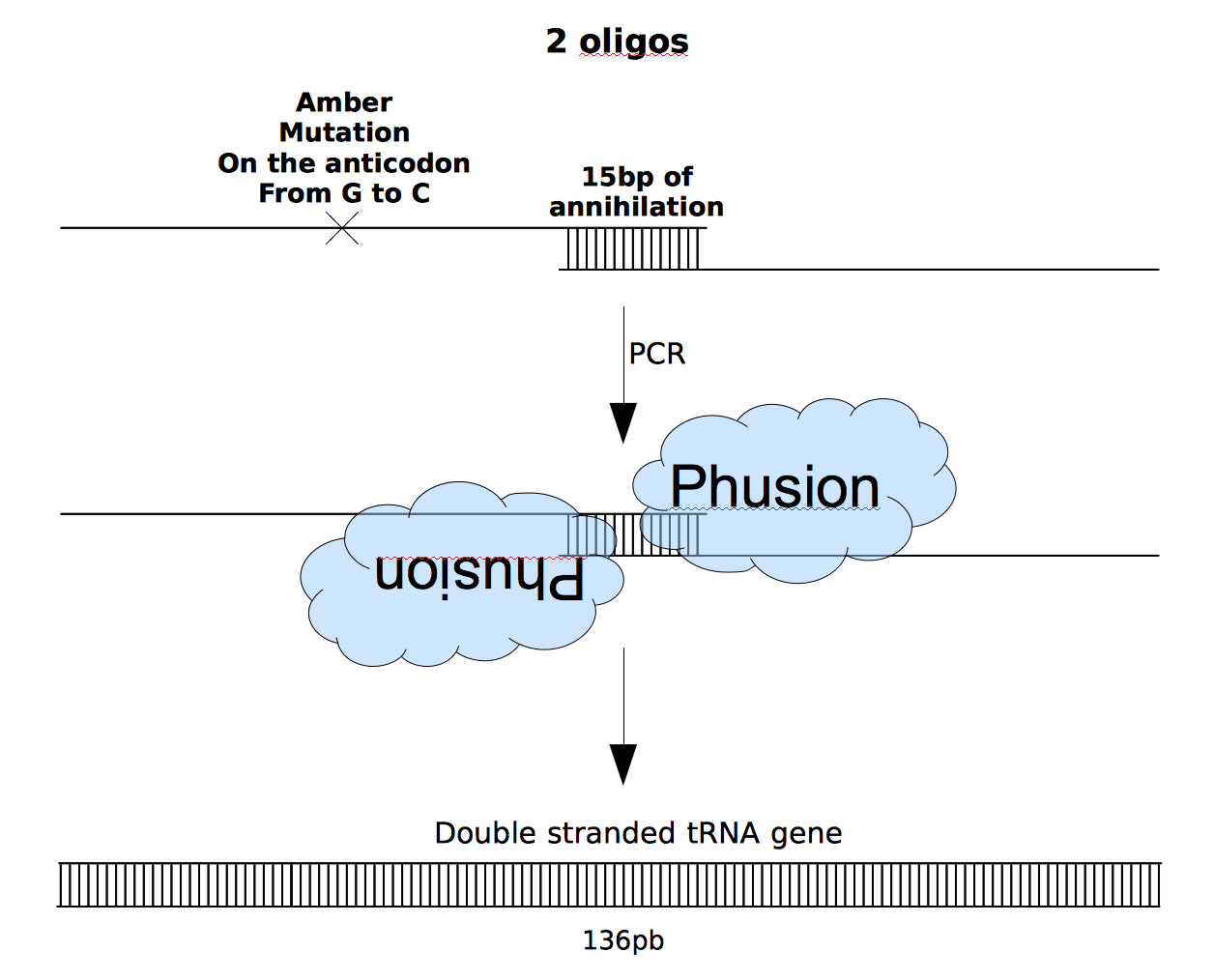
To synthetise the tRNA we thought about using primers and PCR. Indeed the length of it with the biobricks suffix and preffix is only 136bp.
Our idea was :
Therefore we searched for a protocole that would take into account the fact that the melting temperature is increasing with elongation but also the fact that two polymerase will probably not fix at the same moment because 15bp is not long enough to allow them to fix together. Before talking with our supervisors about this experiment, we decided with Cyrille and the advisors of a protocole with cycles and doing a double touchdown PCR.
Our idea :
23 cycles with an anhiling temperature x that would vary along the PCR (touchdown):
- 2mn at 98°C
- 2mn at x °C
- 2mn at 72°C (elongation temperature of the high fidelity Phusion polymerase we used)
For the first touchdown 3 cycles for each temperature :
55 ; 53 ; 51 ; 49 ; 47
For the second touchdown 2 cycles for each temperature :
76 ; 78 ; 79 ; 80
The mix according to Fermentas had to be :
- 10µL Phusion Buffer
- 1µL dNTP (10mM)
- 1.5µL DMSO (tRNAS have big secondary structures)
- yµL of Forward primer in order to have 0.5µM in the final mix
- yµL of Reverse primer in order to have 0.5µM in the final mix
- 0.5µL of Phusion polymerase (Fermentas)
 "
"