Team:Panama/project
From 2011.igem.org
(Difference between revisions)
Ernestopro (Talk | contribs) |
|||
| Line 2: | Line 2: | ||
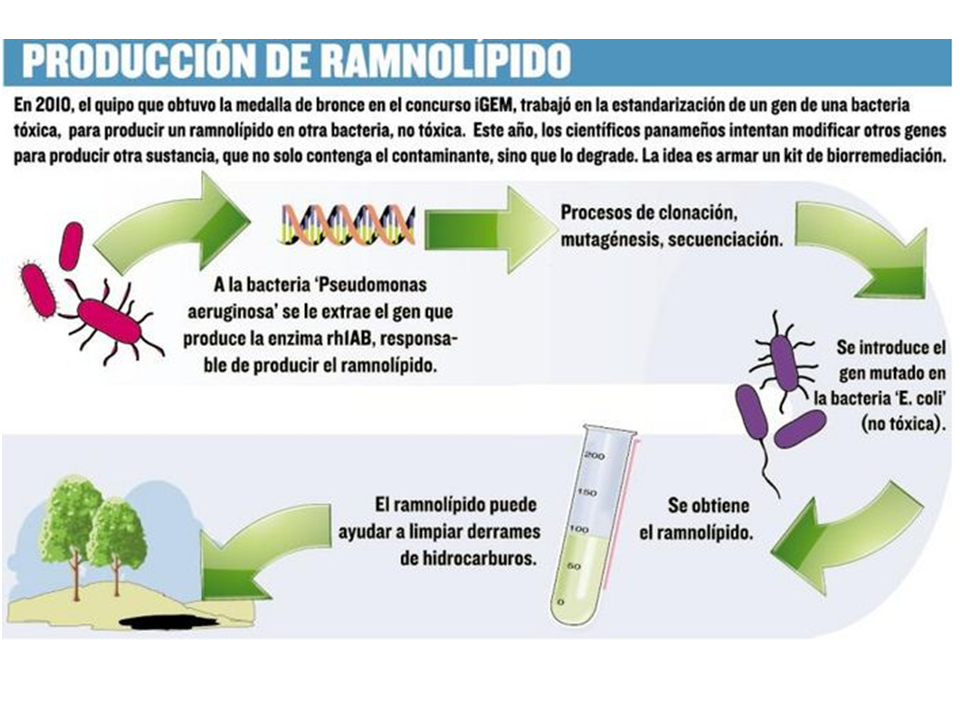
The iGEM Panama Team 2011 is proud to present the second phase of our project '''E. coli based-factory for bio-surfactant production''' or that is what we think so because inconvenients show on. On iGEM 2010 we runned the first phase of the project, entitled: '''Standardization of the Rhamnosiltransferase 1 gene complex (rhlAB) into a Biobrick-friendly for rhamnolipid production in E. coli.''' In iGEM 2010, we build the first panamenian and Central American BioBrick, the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K424018 rhlAB_BB]. The rhamnosyltransferase 1 complex (or, rhlAB) is a naturally occurring gene in ''Pseudomonas aeruginosa'' which synthesizes rhamnolipids, a useful biosurfactant with many applications, for example, it is used by the bio-industries in bioremediation products. The last year, we had been able to extract the genomic DNA of ''P. aeruginosa'' and specially the gene sequence of the rhlAB gene, to make it compatible with Assembly Standard 10 and to put it in, into an ''Escherichia coli'' for standardized rhamnolipid production, by the means of synthetic biology based on standardized biological parts. So, we take advances in genetic engineering as synthetic biology, as viable solution to oil pollution cleanup. But for do this; we need to design and build an expression platform that works into our modified ''E.coli''. For this, We choose four standardized biological parts from the Registry, those parts are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005 Promoter T7], [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0034 Ribosomal Binding Site (RBS)], [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0040 GFP Reporter] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0015 Double Terminator]; Were selected using rationale from molecular biology and genetic engineering to enable us to build up rationally-engineered, our expression platform. For the iGEM 2011, we will integrate and assembly those parts (P+RBS+GFP+T) along with our BioBrick part using the [http://openwetware.org/wiki/Amplified_Insert_Assembly Amplified Insert Assembly method], to bring up an expression device [http://partsregistry.org/wiki/index.php?title=Part:BBa_K424017 (P+RBS+rhlAB_BB+GFP+T)] for rhamnolipid production in ''E. coli''. But reality shows us a different challenge on the way. Our last year BioBrick the rhamnosyltransferace BioBrick appears to be denaturalized from the plasmid, thus the insertion was unsuccesful for long term. Now we will re-design and re-assembly our BioBrick part [http://partsregistry.org/Part:BBa_K653000:Design RhlAB_BBa_K653000] in order to acheive '''Quality'''. | The iGEM Panama Team 2011 is proud to present the second phase of our project '''E. coli based-factory for bio-surfactant production''' or that is what we think so because inconvenients show on. On iGEM 2010 we runned the first phase of the project, entitled: '''Standardization of the Rhamnosiltransferase 1 gene complex (rhlAB) into a Biobrick-friendly for rhamnolipid production in E. coli.''' In iGEM 2010, we build the first panamenian and Central American BioBrick, the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K424018 rhlAB_BB]. The rhamnosyltransferase 1 complex (or, rhlAB) is a naturally occurring gene in ''Pseudomonas aeruginosa'' which synthesizes rhamnolipids, a useful biosurfactant with many applications, for example, it is used by the bio-industries in bioremediation products. The last year, we had been able to extract the genomic DNA of ''P. aeruginosa'' and specially the gene sequence of the rhlAB gene, to make it compatible with Assembly Standard 10 and to put it in, into an ''Escherichia coli'' for standardized rhamnolipid production, by the means of synthetic biology based on standardized biological parts. So, we take advances in genetic engineering as synthetic biology, as viable solution to oil pollution cleanup. But for do this; we need to design and build an expression platform that works into our modified ''E.coli''. For this, We choose four standardized biological parts from the Registry, those parts are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005 Promoter T7], [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0034 Ribosomal Binding Site (RBS)], [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0040 GFP Reporter] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0015 Double Terminator]; Were selected using rationale from molecular biology and genetic engineering to enable us to build up rationally-engineered, our expression platform. For the iGEM 2011, we will integrate and assembly those parts (P+RBS+GFP+T) along with our BioBrick part using the [http://openwetware.org/wiki/Amplified_Insert_Assembly Amplified Insert Assembly method], to bring up an expression device [http://partsregistry.org/wiki/index.php?title=Part:BBa_K424017 (P+RBS+rhlAB_BB+GFP+T)] for rhamnolipid production in ''E. coli''. But reality shows us a different challenge on the way. Our last year BioBrick the rhamnosyltransferace BioBrick appears to be denaturalized from the plasmid, thus the insertion was unsuccesful for long term. Now we will re-design and re-assembly our BioBrick part [http://partsregistry.org/Part:BBa_K653000:Design RhlAB_BBa_K653000] in order to acheive '''Quality'''. | ||
| + | At the end of all the assemblies, we visualize for the future an application like this: | ||
| + | |||
| + | [[File:RhlABproduction.png|centre|600px]] | ||
Now the follow links will show you part of how we work in a funny way on the project! | Now the follow links will show you part of how we work in a funny way on the project! | ||
Latest revision as of 05:23, 29 September 2011
Overall Project
The iGEM Panama Team 2011 is proud to present the second phase of our project E. coli based-factory for bio-surfactant production or that is what we think so because inconvenients show on. On iGEM 2010 we runned the first phase of the project, entitled: Standardization of the Rhamnosiltransferase 1 gene complex (rhlAB) into a Biobrick-friendly for rhamnolipid production in E. coli. In iGEM 2010, we build the first panamenian and Central American BioBrick, the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K424018 rhlAB_BB]. The rhamnosyltransferase 1 complex (or, rhlAB) is a naturally occurring gene in Pseudomonas aeruginosa which synthesizes rhamnolipids, a useful biosurfactant with many applications, for example, it is used by the bio-industries in bioremediation products. The last year, we had been able to extract the genomic DNA of P. aeruginosa and specially the gene sequence of the rhlAB gene, to make it compatible with Assembly Standard 10 and to put it in, into an Escherichia coli for standardized rhamnolipid production, by the means of synthetic biology based on standardized biological parts. So, we take advances in genetic engineering as synthetic biology, as viable solution to oil pollution cleanup. But for do this; we need to design and build an expression platform that works into our modified E.coli. For this, We choose four standardized biological parts from the Registry, those parts are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005 Promoter T7], [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0034 Ribosomal Binding Site (RBS)], [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0040 GFP Reporter] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0015 Double Terminator]; Were selected using rationale from molecular biology and genetic engineering to enable us to build up rationally-engineered, our expression platform. For the iGEM 2011, we will integrate and assembly those parts (P+RBS+GFP+T) along with our BioBrick part using the [http://openwetware.org/wiki/Amplified_Insert_Assembly Amplified Insert Assembly method], to bring up an expression device [http://partsregistry.org/wiki/index.php?title=Part:BBa_K424017 (P+RBS+rhlAB_BB+GFP+T)] for rhamnolipid production in E. coli. But reality shows us a different challenge on the way. Our last year BioBrick the rhamnosyltransferace BioBrick appears to be denaturalized from the plasmid, thus the insertion was unsuccesful for long term. Now we will re-design and re-assembly our BioBrick part [http://partsregistry.org/Part:BBa_K653000:Design RhlAB_BBa_K653000] in order to acheive Quality.At the end of all the assemblies, we visualize for the future an application like this:
Now the follow links will show you part of how we work in a funny way on the project!
 "
"