Team:Northwestern/Project/Introduction
From 2011.igem.org
Helenmelon (Talk | contribs) |
Helenmelon (Talk | contribs) |
||
| Line 23: | Line 23: | ||
'''Figure 2:''' Genetic circuit for ''E. coli'', incorporating several elements from ''P. aeruginosa''. | '''Figure 2:''' Genetic circuit for ''E. coli'', incorporating several elements from ''P. aeruginosa''. | ||
</DIV></DIV> | </DIV></DIV> | ||
| + | |||
| + | |||
| + | ---------------------- | ||
| + | [[Image:Sponsor_northwestern.jpg]] | ||
| + | [[Image:Sponsor_weinberg.jpg]] | ||
| + | [[Image:Sponsor_mccormick.jpg]] | ||
Revision as of 02:48, 13 July 2011
PROJECT

RESULTS

CONSIDERATIONS

ABOUT US

NOTEBOOK

ATTRIBUTIONS

Introduction
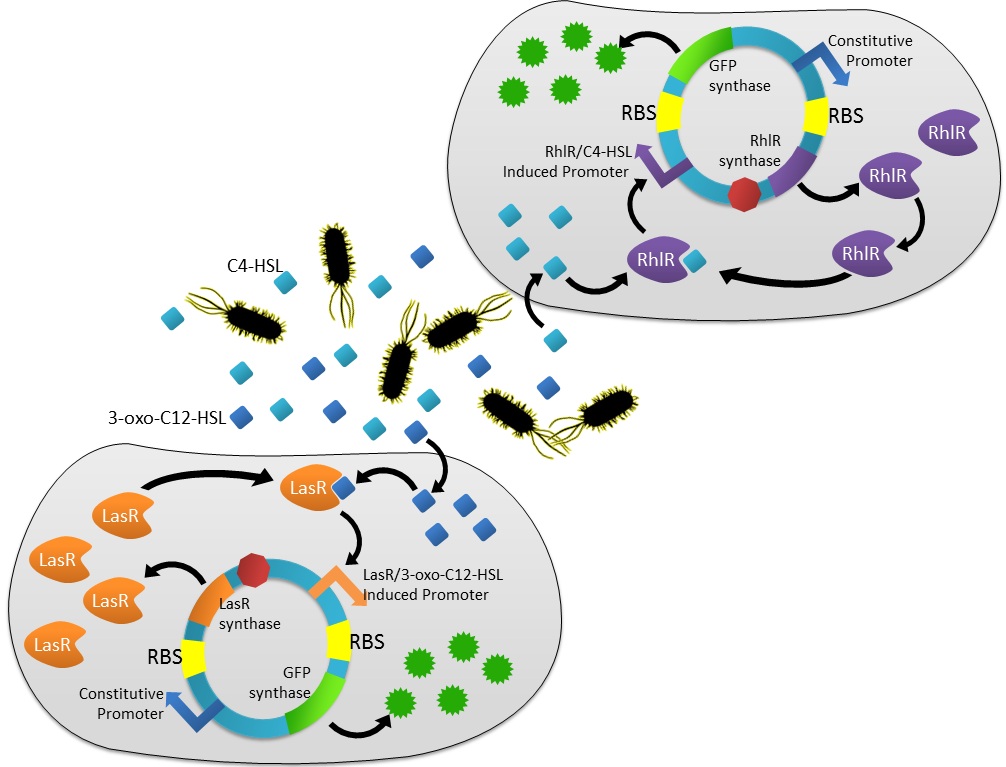
Virulence factors produced by Pseudomonas aeruginosa are controlled by a natural regulatory, hierarchical network consisting of two main systems: the las system and the rhl system. The main components of these systems are the receptor proteins (lasR and rhlR) and the autoinducers (3-oxo-C12-HSL and C4-HSL). The formation of a lasR/3-oxo-C12-HSL complex activates the transcription of several genes, notably rhlR as well as others responsible for biofilm differentiation. The binding of the C4-HSL autoinducer to rhlR further activates genes including the rhlAB operon, which encodes enzymes for the production of rhamnolipids that modulate the motility of P. aeruginosa. Both lasR and rhlR are subject to positive feedback autoregulation through autoinducers. Figure 1 details the existing quorum sensing system inherent to P. aeruginosa.
Our project goal is to utilize several genes from this existing system, specifically to introduce las and rhl receptors into E. coli in order to sense the presence of 3-oxo-C12-HSL and/or C4-HSL, thereby detecting P. Aeruginosa. Figure 2 illustrates the genetic circuit that we are attempting to incorporate into E. coli.
Figure 2: Genetic circuit for E. coli, incorporating several elements from P. aeruginosa.
 "
"