Team:NYMU-Taipei/synthoprime
From 2011.igem.org


Contents |
Introduction
- SynthoPrime – A Synthetic Biology Primer Design Tool for Genomic Parts Creation
 ]
]
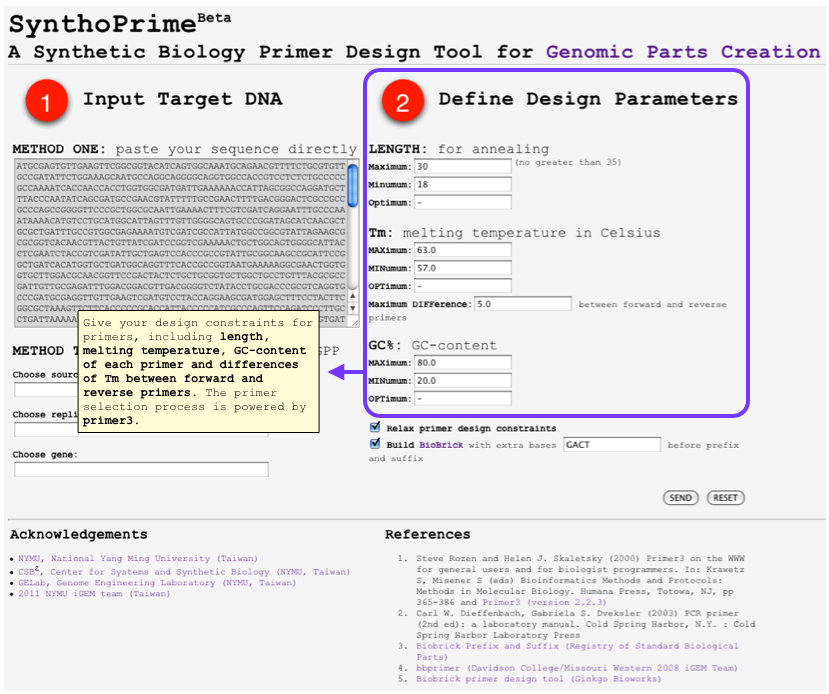
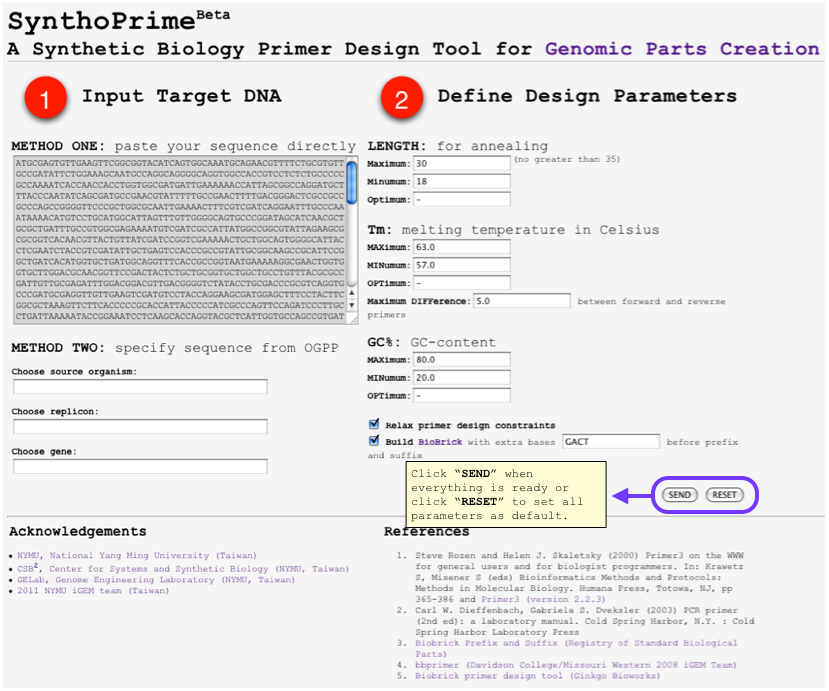
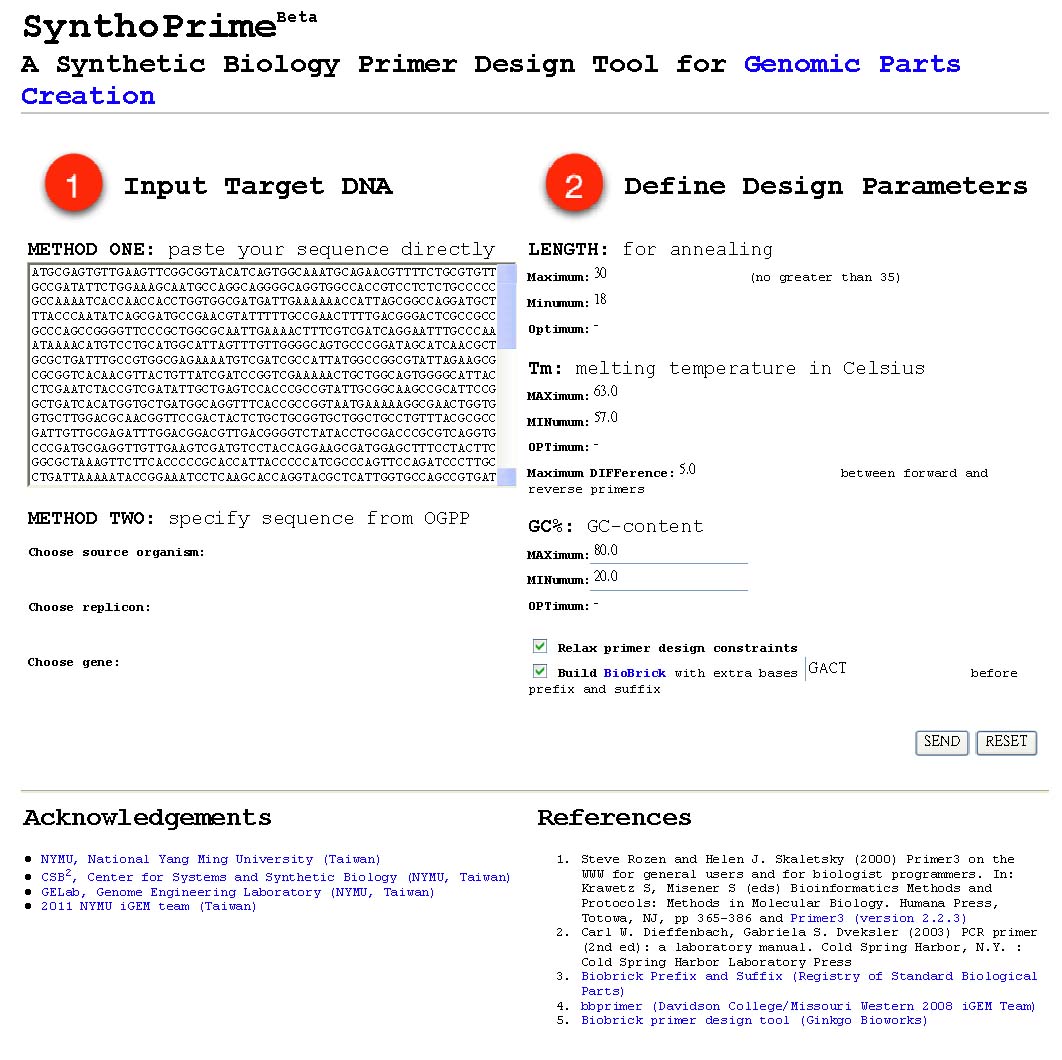
- [http://120.126.44.58/SynthoPrime/ SynthoPrime] algorithm is developed to provide the access of all the available genomic parts (biological parts from sequenced genomes) for the specific needs of primer design in synthetic biology communities. It provides an automated ONE-STEP primer design service of BioBricks for iGEM. Near 1,400 bacteria and their 2,551 replicons (chromosomes and plasmids) are extracted from NCBI genome database by BioPerl package as genomic templates in our service. The jQuery Javascript library is used for interactive web user interface design to help synthetic biologists to access genomic parts from the sequenced genome templates for primer design. Our online service aims to provide a one-step primer design tool and sequenced genomes as templates to convert sequenced genomes into biological parts with quantitative measures for synthetic biology community. [http://120.126.44.58/SynthoPrime/ SynthoPrime] is freely available for everyone to try out.
Features
- By using [http://120.126.44.58/SynthoPrime/ SynthoPrime] program, you can design your own biobrick primers by selecting genes that you need with quantitative measures, including primer length, melting temperature and GC content.
- You can access any genes from sequenced organism and you can also directly input the total sequence of that specific gene if you’ve already got the sequence.
- In the output, you will receive multiple results of optimal primers for your gene and associated information of each candidate, this includes melting temperature, GC content, self annealing score, estimated price etc.
- This Program provides an automatic ONE-STEP primer design service of biobrick for iGEM communty to access all available genomic parts.
HOWTO:User Inputs
HOWTO:Output Reading
References and Feedback
 "
"