Team:NYMU-Taipei/synthoprime
From 2011.igem.org
(→HOWTO: User Inputs) |
|||
| (30 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NYMU-Taipei/Templates/Header/menubar}} | {{:Team:NYMU-Taipei/Templates/Header/menubar}} | ||
| - | |||
| - | |||
| - | + | ==<font size=5><font color=crimson>'''Introduction'''</font></font>== | |
| - | + | ||
| - | <font size= | + | |
| - | + | ||
| - | + | <font size=4>[http://120.126.44.58/SynthoPrime/ SynthoPrime] – A Synthetic Biology Primer Design Tool for Genomic Parts Creation</font> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| + | <center>[[File:SynthoPrime.jpg|700px|link=http://120.126.44.58/SynthoPrime/|caption]]</center> | ||
| + | <font size=3> | ||
| + | <font color=red>[http://120.126.44.58/SynthoPrime/ SynthoPrime]</font> algorithm is developed to provide the access of all the available genomic parts (biological parts from sequenced genomes) for the specific needs of primer design in synthetic biology communities. <font color=medium blue>It provides an automated ONE-STEP primer design service</font> of BioBricks for iGEM. <font color=medium blue>Near 1,400 bacteria and their 2,551 replicons (chromosomes and plasmids) are extracted</font> from NCBI genome database by BioPerl package as genomic templates in our service. The jQuery Javascript library is used for interactive web user interface design to help synthetic biologists to access genomic parts from the sequenced genome templates for primer design. <font color=medium blue>Our online service aims to provide a one-step primer design tool and sequenced genomes as templates to convert sequenced genomes into biological parts with quantitative measures for synthetic biology community.</font> [http://120.126.44.58/SynthoPrime/ SynthoPrime] is freely available for everyone to try out. | ||
| - | + | ==<font size=5><font color=crimson>'''HOWTO: User Inputs'''</font></font>== | |
| - | + | <center> | |
{| | {| | ||
| valign=top| | | valign=top| | ||
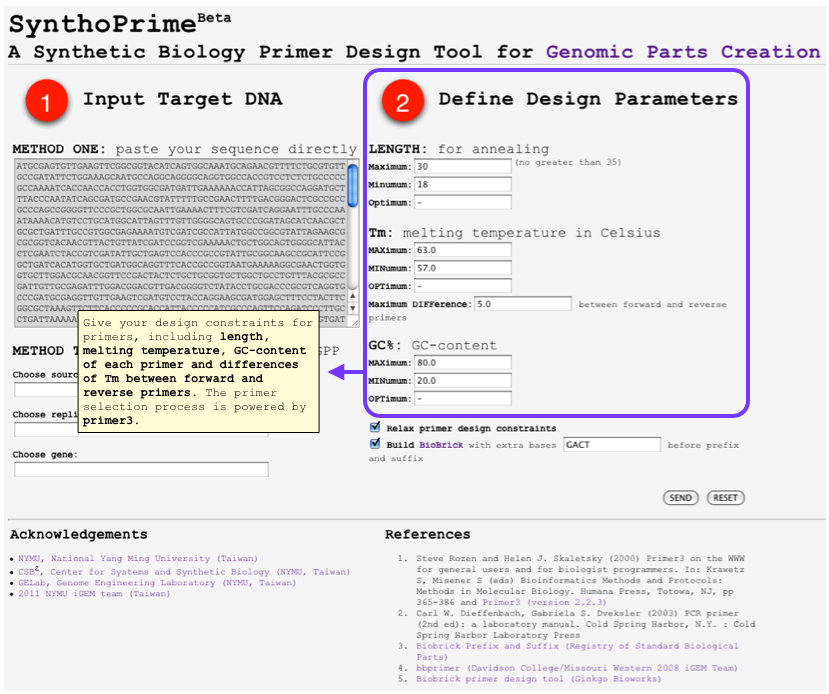
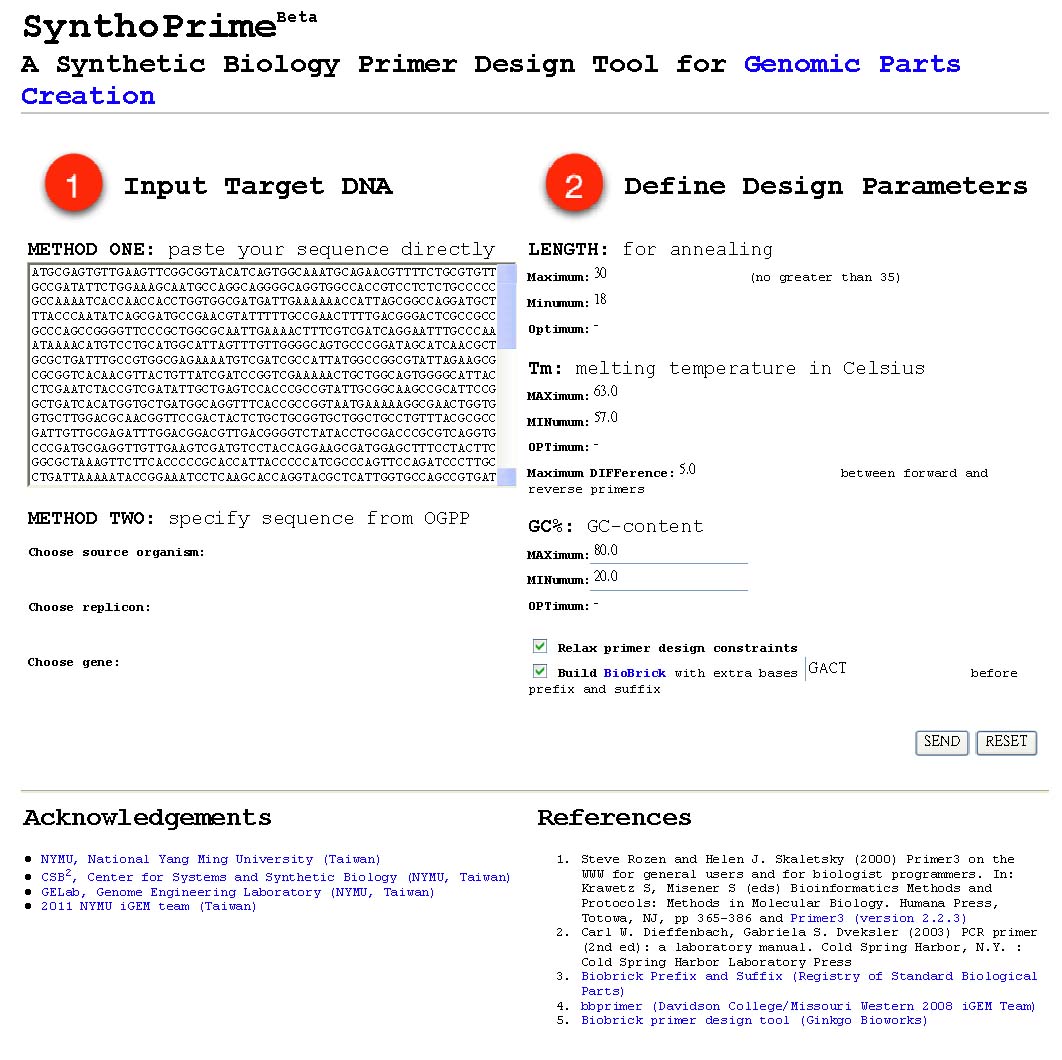
[[File:SynthoPrime manual p1.png|thumb|300px| '''STEP 1:''' Offer your sequence to be ‘biobricked’ by direct pasting your DNA or specifying by names of organism, replicon and gene.]] | [[File:SynthoPrime manual p1.png|thumb|300px| '''STEP 1:''' Offer your sequence to be ‘biobricked’ by direct pasting your DNA or specifying by names of organism, replicon and gene.]] | ||
| valign=top| | | valign=top| | ||
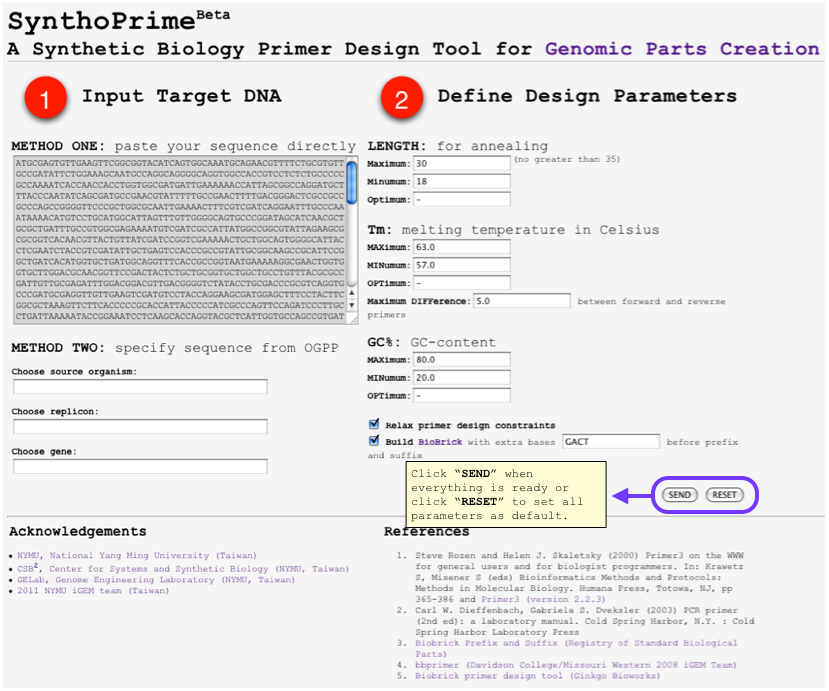
| - | [[File:SynthoPrime manual p2.png|thumb|300px| ''' | + | [[File:SynthoPrime manual p2.png|thumb|300px| '''STEP 2:''' Give your design constraints for primers, including length, melting temperature, GC-content of each primer and differences of Tm between forward and reverse primers. The primer selection process is powered by primer3.]] |
|- | |- | ||
| valign=top| | | valign=top| | ||
| Line 34: | Line 26: | ||
|- | |- | ||
|} | |} | ||
| + | </center> | ||
| - | + | <center><html><object width="640" height="360"><param name="movie" value="http://www.youtube.com/v/6n_l3rCVS_g?version=3&hl=zh_TW"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/6n_l3rCVS_g?version=3&hl=zh_TW" type="application/x-shockwave-flash" width="640" height="360" allowscriptaccess="always" allowfullscreen="true"></embed></object></html></center> | |
| + | ==<font size=5><font color=crimson>'''HOWTO: Output Reading'''</font></font>== | ||
| + | <center> | ||
{| | {| | ||
| valign=top| | | valign=top| | ||
| Line 45: | Line 40: | ||
|- | |- | ||
|} | |} | ||
| + | </center> | ||
| - | + | <center><html><object width="640" height="480"><param name="movie" value="http://www.youtube.com/v/DTSXX2i7yjM?version=3&hl=zh_TW"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/DTSXX2i7yjM?version=3&hl=zh_TW" type="application/x-shockwave-flash" width="640" height="480" allowscriptaccess="always" allowfullscreen="true"></embed></object></html></center> | |
| + | ==<font size=5><font color=crimson>'''References and Feedback '''</font></font>== | ||
| + | <center> | ||
{| | {| | ||
| valign=top| | | valign=top| | ||
| - | [[File:SynthoPrime manual p7.png|thumb|630px| We would be more than happy to receive your comments and suggestions.]] | + | [[File:SynthoPrime manual p7.png|thumb|630px| '''We would be more than happy to receive your comments and suggestions.''']] |
|- | |- | ||
|} | |} | ||
| - | </ | + | </center> |
Latest revision as of 01:13, 29 October 2011

Contents |
Introduction
[http://120.126.44.58/SynthoPrime/ SynthoPrime] – A Synthetic Biology Primer Design Tool for Genomic Parts Creation
[http://120.126.44.58/SynthoPrime/ SynthoPrime] algorithm is developed to provide the access of all the available genomic parts (biological parts from sequenced genomes) for the specific needs of primer design in synthetic biology communities. It provides an automated ONE-STEP primer design service of BioBricks for iGEM. Near 1,400 bacteria and their 2,551 replicons (chromosomes and plasmids) are extracted from NCBI genome database by BioPerl package as genomic templates in our service. The jQuery Javascript library is used for interactive web user interface design to help synthetic biologists to access genomic parts from the sequenced genome templates for primer design. Our online service aims to provide a one-step primer design tool and sequenced genomes as templates to convert sequenced genomes into biological parts with quantitative measures for synthetic biology community. [http://120.126.44.58/SynthoPrime/ SynthoPrime] is freely available for everyone to try out.
 "
"