Team:NYC Software/Deinococcus/Previous Studies
From 2011.igem.org
(Difference between revisions)
m |
|||
| Line 20: | Line 20: | ||
<div id="container"> | <div id="container"> | ||
<br><br> | <br><br> | ||
| + | <h4>What we have learned about the Deinococcus Genera</h4> | ||
| + | <h4>Different Phylogenetic Methods Yield Different Clades</h4> | ||
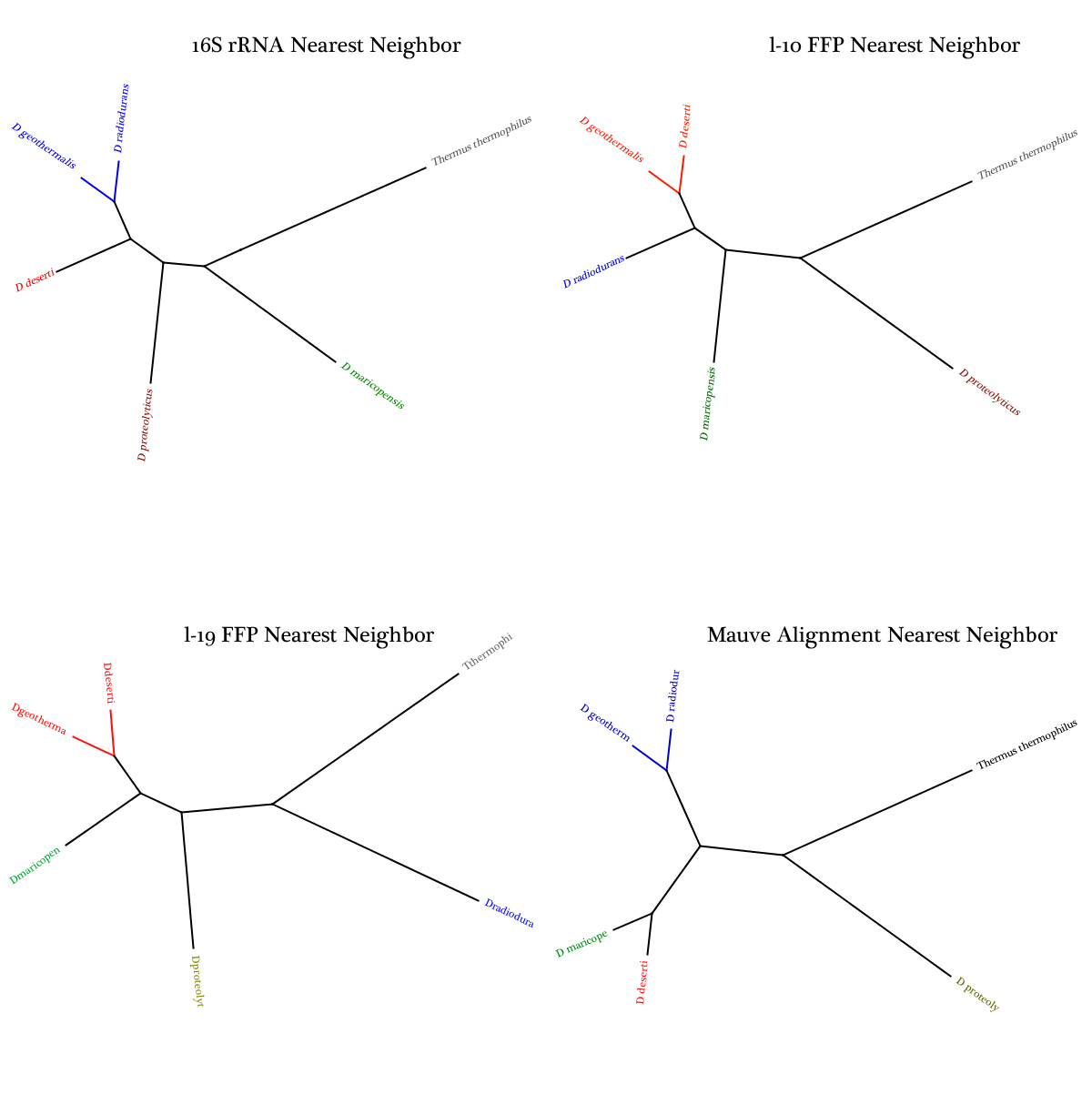
| - | + | We extrapolated the evolutionary relationships between the species using 1) Feature Frequency Profile comparisons, 2) 16S rRNA alignments, and 3) Mauve genome alignment, but each method yielded an alternative result. | |
| - | + | <br>By sequencing more species in this family, we hypothesize that Inclusion of a larger set of sequenced species in the genera may elucidate new and more reliable information about their origins and cladistics. | |
| - | + | ||
<p> | <p> | ||
| - | <strong>Disparate Trees Resulting From Too Little Information</strong> | + | <strong>Disparate Trees Resulting From Too Little Genetic Information</strong> |
Latest revision as of 00:34, 29 September 2011

What we have learned about the Deinococcus Genera
Different Phylogenetic Methods Yield Different Clades
We extrapolated the evolutionary relationships between the species using 1) Feature Frequency Profile comparisons, 2) 16S rRNA alignments, and 3) Mauve genome alignment, but each method yielded an alternative result.By sequencing more species in this family, we hypothesize that Inclusion of a larger set of sequenced species in the genera may elucidate new and more reliable information about their origins and cladistics.
Disparate Trees Resulting From Too Little Genetic Information
 "
"