Team:Macquarie Australia/Project
From 2011.igem.org
(→The Phytochromes) |
(→Heme Oxygenase 1) |
||
| Line 155: | Line 155: | ||
==Heme Oxygenase 1== | ==Heme Oxygenase 1== | ||
| - | The PCR optimisation of the RBS-coupled heme oxygenase 1 coding sequence was successful from the start (see notebook), and much more effort was put into the construction of the HO-BioBrick, and the assembly with a T7 promoter (BBa_I719005 and BBa_I712074). | + | The PCR optimisation of the RBS-coupled heme oxygenase 1 coding sequence was successful from the start (see [https://2011.igem.org/Team:Macquarie_Australia/Notebook#12-19.2F8.2F2011 notebook]), and much more effort was put into the construction of the HO-BioBrick, and the assembly with a T7 promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005 BBa_I719005] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I712074 BBa_I712074]). |
| - | After the initial failure of the ligation of the HO-PCR product with the plasmid backbone, and the lack of specificity of the colony PCR (see notebook), the BioBrick construction pipeline was adopted, using a new batch of alkaline phosphatase treated backbone, and the ''Xba''I restriction digest as a screening method. Despite getting only 1 colony from the transformation, the single colony proved to be right on target, giving us our first successful BioBrick. | + | After the initial failure of the ligation of the HO-PCR product with the plasmid backbone, and the lack of specificity of the colony PCR (see [https://2011.igem.org/Team:Macquarie_Australia/Notebook#2.2F9.2F2011 notebook]), the BioBrick construction pipeline was adopted, using a new batch of alkaline phosphatase treated backbone, and the ''Xba''I restriction digest as a screening method. Despite getting only 1 colony from the transformation, the single colony proved to be right on target, giving us our first successful BioBrick. |
[[File:Results_-_HO_screen.jpg|150px]][[File:Results_-_HO-bb_confirmation.jpg|150px]] | [[File:Results_-_HO_screen.jpg|150px]][[File:Results_-_HO-bb_confirmation.jpg|150px]] | ||
| Line 163: | Line 163: | ||
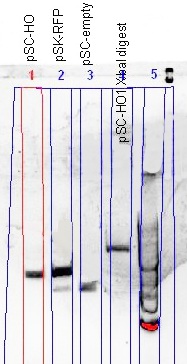
(Left) The comparison of the uncut plasmid size (pSC-HO) with the RFP-plasmid (pSC-RFP) and the re-ligated plasmid (pSC-empty) showed a successful insertion of the HO gene; smaller than RFP-plasmid, but larger than the re-ligated plasmid. The ''Xba''I digest gave prove of the gene orientation; a plasmid able to be digested with ''Xba''I. | (Left) The comparison of the uncut plasmid size (pSC-HO) with the RFP-plasmid (pSC-RFP) and the re-ligated plasmid (pSC-empty) showed a successful insertion of the HO gene; smaller than RFP-plasmid, but larger than the re-ligated plasmid. The ''Xba''I digest gave prove of the gene orientation; a plasmid able to be digested with ''Xba''I. | ||
| - | (Right) Confirmation of the HO-BioBrick (BBa_K646000) was performed, using both PCR and a restriction digest using ''Xba''I and ''Pst''I. Although the gel was not well resolved, the similarity of the PCR product size when compared with the original template, and that a gene fragment was cleaved out of the HO-BioBrick, was good enough to conclude that the HO-BioBrick construction was successful. | + | (Right) Confirmation of the HO-BioBrick ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K646000 BBa_K646000]) was performed, using both PCR and a restriction digest using ''Xba''I and ''Pst''I. Although the gel was not well resolved, the similarity of the PCR product size when compared with the original template, and that a gene fragment was cleaved out of the HO-BioBrick, was good enough to conclude that the HO-BioBrick construction was successful. |
Revision as of 10:38, 3 October 2011
Contents |
Our project
Background
Phytochromes were first discovered in plant and were found to be associated with the plants photoperception. The phytochrome proteins act as light sensors by absorbing red/far red light, with some their range can be between the UV-A to far-red regions. Their main function was found to mediate various time-dependant events such as seed germination, seedling de-etiolation and the generation of flowers.
Bacteriophytochromes also act as photoreceptors absorbing in the red to far-red light, they mediate sensory responses to their light environment, the alignment of their circadian system's can function as a quorum-sensing network. Bacteriophytochromes can act as protein kinases sensitive to the light which are able to modify or regulate various components in the cell, such as protein activity and the synthesis of the light harvesting complex via the promotion of transcription factors. They control genes associated with various rhythm-dependant proteins at the transcription level by regulation of transcription factors to the promoter regions depending on their binding affinity. By doing so, they create what is called, a “light switch”. Techniques can be used to temporarily inactivate specific proteins by using a photolabile protecting group; this is referred to as “caging”. The light switch bacteriophytochrome that is utilised by Agrobacterium and Deinococcus has a domain structure as follows, PAS-GAF-PHY-HisKinase and is able to cause a shift in conformation of the chromophore complex according to the wavelength absorbed.
For a light switch to be self-sufficient and fully functional, two main components are required, a hemeoxygenase and a bacteriophytochrome coupled with ribosome binding sites also known as the Shine-Dalgarno sequence. The hemeoxygenase is required for the degradation of the porphyrin heme to form the linear tetrapyrrole biliverdin which is then able to autocatalytically bind to the bacteriophytochrome on the GAF domain of the bacteriophytochrome.
This forms the chromophore complex producing a colour according to the ratio of red to far-red light absorbed. Various processes are influenced by phytochromes through photoconversion between two isomers, a phytochrome red biologically inactive form (Pr) that absorbs red light and a phytochrome far red active form which absorbs far-red light. Pr expresses a blue colour that is able to isomerise to a green colour when absorption of far-red light is greater.
The bacteriophytochrome vector will be transformed into E.coli competent BL21 (DE3) cells. However E.coli does not contain a hemeoxygenase gene which is why it must be incorporated into the genetically engineered operon along with a T7 promoter/terminator and the bacteriophytochrome genes from Deinococcus and Agrobacterium.
Aims
The objective in this project is to build and characterise a biological light switch in E. coli. This will involve construction of heme-oxygenase and bacteriophytochrome BioBrick parts. In 2010 the Macquarie Team cloned bacteriophytochrome from two sources. They showed that when one was expressed, it was functionally assembled when incubated with exogenous biliverdin; able to elicit a colour change when excited with far-red light. However, the part created is not directly usable as a BioBrick as it contains an internal EcoRI site (Deinococcus radiodurans phytochrome) and 2 PstI sites (Agrobacterium tumefaciens phytochrome). As biliverdin is not native to E. coli, the addition of heme oxygenase is required for the synthesis of bilivedin, enabling the self-assembly of the light switch. The 2010 Macquarie Team had managed to combine a ribosome binding site (RBS) to all 3 proteins of interest.
Hence, the aims of the team this year are to complete the light switch construction:
1. Assemble 3 fully functional BioBricks which are functionally expressed in E.coli.
2. Remove the restriction sites from the bacteriophytochrome which are incompatible with BioBrick assembly.
3. Assemble an operon consisting of the heme-oxygenase and bacteriophytochrome BioBrick parts.
4. Optimise the gene expression from the operon such that the bacteriophytochrome light switch works without requiring the addition of biliverdin.
The Experiments
Primer Design
Primer Sequence Melting temp (°C) BB-FWD 5'- GAA TTC GCG GCC GCT TCT AGA -3' 59.7 BB-REV 5'- CTG CAG CGG CCG CTA CTA GTA -3' 61 T7-BB-FWD 5'- GAA TTC GCG GCC GCT TCT AGA TCT CGA TCC CGC G -3' 69.4 T7-BB-REV 5'- CTG CAG CGG CCG CTA CTA GTA GAG GGG AAT TGT TAT -3' 66.5 HO1-BB-FWD 5'- GCG GCC GCT TCT AGA CTT TAA GAA GGA GAT ATA C -3' 61.9 HO1-BB-REV 5'- GCG GCC GCT ACT AGT ACT TTC GGG CTT TGT TAG C -3' 67 DR-BB-FWD 5'- TCG CGG CCG CTT CTA GAA GGA GGG CTG CTA TGA GC -3' 71 DR-BB-REV 5'- GCG GCC GCT ACT AGT ATC AGG CAT CGG CGG CTC CCG G -3' 68.7 DR-M-FWD 5'- TTG CTG ATT CTG GAG TTC GAG CCG ACG GAG -3' 63.5 DR-M-REV 5'- CTC CGT CGG CTC GAA CTC CAG AAT CAG CAA -3' 61.7 AT-BB-FWD 5'- CGC GGC CGC TTC TAG AGA TTA GGA GGG CTG CTA TGA G -3' 69.1 AT-BB-REV 5'- GCG GCC GCT ACT AGT AGA TTT CAG GCA ATT TTT TCC -3' 64.3 AT-M1-FWD 5'- GTC CCT GCA TTA TCT TCA GAT GAT ATC AGA -3' 53.8 AT-M1-REV 5'- TCT GAT ATC ATC TGA AGA TAA TGC AGG GAC -3' 55.7 AT-M2-FWD 5'- GTG TTT CAG AGG CTT CAG CGG GTG GAG GA -3' 63.9 AT-M2-REV 5'- TCC TCC ACC CGC TGA AGC CTC TGA AAC AC -3' 64.5 BB - BioBrick compatible, M - Mutagenic primer (Italics - base mutation), Bold - Restriction sites
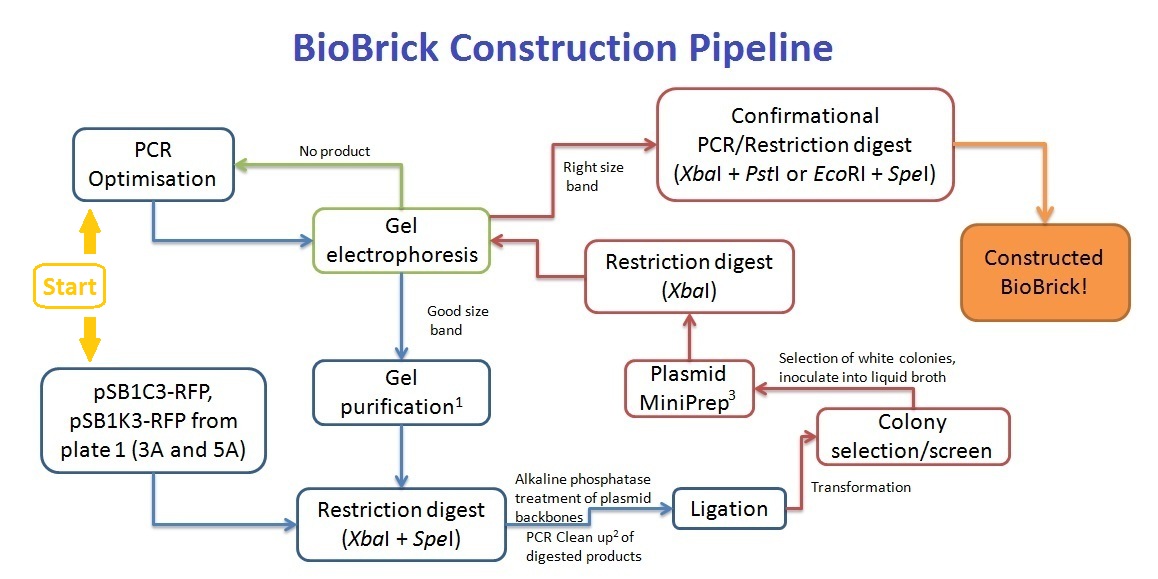
Primers were designed such that the amplicon would consists of at least the XbaI site or SpeI site; restricted by the length of the primers. As such, an EcoRI or PstI restriction digest would not work for our BioBrick construction, resulting in the XbaI and SpeI digestion for the ligation. This then gives rise in the problem where the inserted coding sequence would be in the wrong orientation; SpeI ligating with XbaI instead. To solve this problem, we've decided to screen at least 10 colonies that grow on the antibiotic resistant plates, and perform an XbaI digest to screen for inserts that are in the right orientation. Colony PCR was trialled, but due to the lack of difference between the right and wrong orientation of inserts (only 2 single base difference), the screening method was abandoned (see notebook).
BioBrick Construction
1 - Qiagen Gel extraction kit, 2 - Sigma PCR clean up kit, 3 - Qiagen Spin Miniprep kit
Along with the PCR clean up of the pre-ligation products, the cut plasmid backbone was separated on a gel, and gel purified to remove any red fluorescent protein gene products.
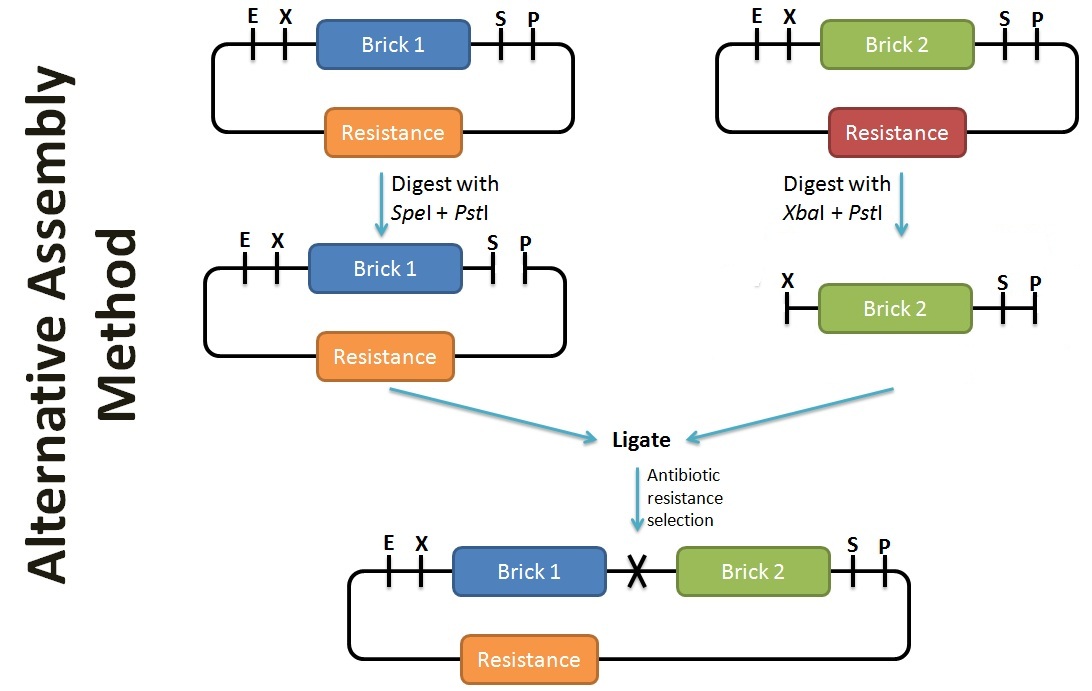
Alternative Assembly Method
In contrast to the assembly protocol as proposed by iGEM, we've adopted a slightly different protocol. This can be applied when the antibiotic backbone of one of the parts remains the same (such as preparation for submission), while inserting the desired part. The screening of successful assembly can either be by protein expression (eg. T7 promoter part + GFP = fluoresce), PCR using BioBrick primers, or by plasmid extraction (successful assembly would have a larger band size). To enhance the efficiency of the assembly, alkaline phophatase treatment should be performed on the plasmid with the selection resistance to minimise re-ligation.
Results
The Phytochromes
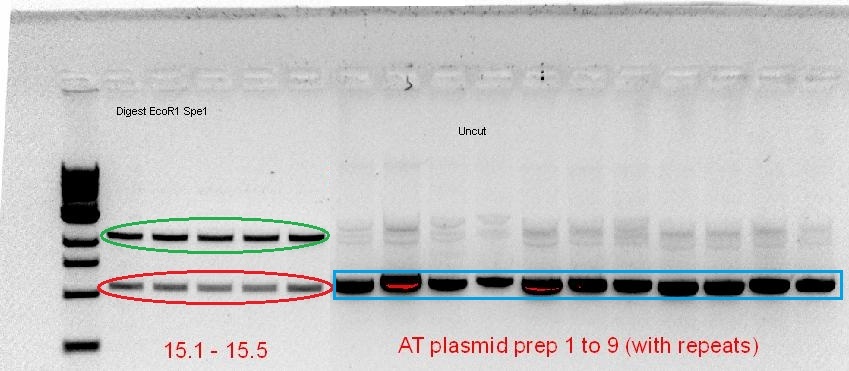
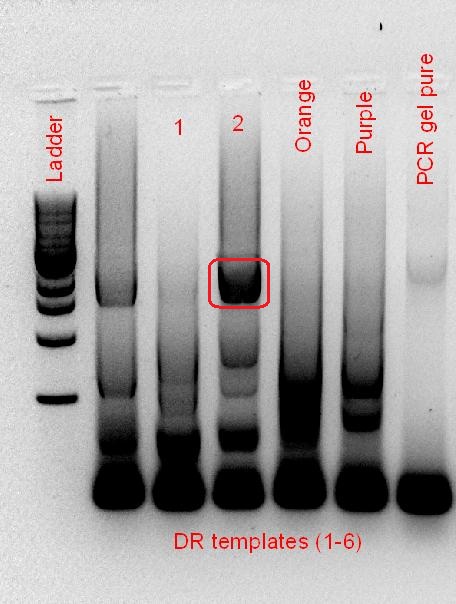
The starting template that we had to work with was a RBS-coupled phytochrome coding sequence containing plasmid (for A. tumefaciens) and PCR product (for D. radiodurans) from the 2010 Macquarie iGEM team. Using the BioBrick compatible primers designed, PCR optimisation for the 2 phytochromes was screened over a range of temperatures, buffer types and cycle conditions (see notebook). When the desired product size was visible on the gel, the band was cut out and gel purified, subsequently following the BioBrick construction pipeline as mentioned. However, after the screening of the purified plasmid from the transformed cells, it seems that the plasmid contained no insert (for AT-Bph) or the wrong insert (for DR-Bph).
Green - plasmid backbone, Red - Size of ~1kb, corresponds with size of Red fluorescent protein, Blue - Size of plasmid show that there is no insert (based on previous experience, see notebook)
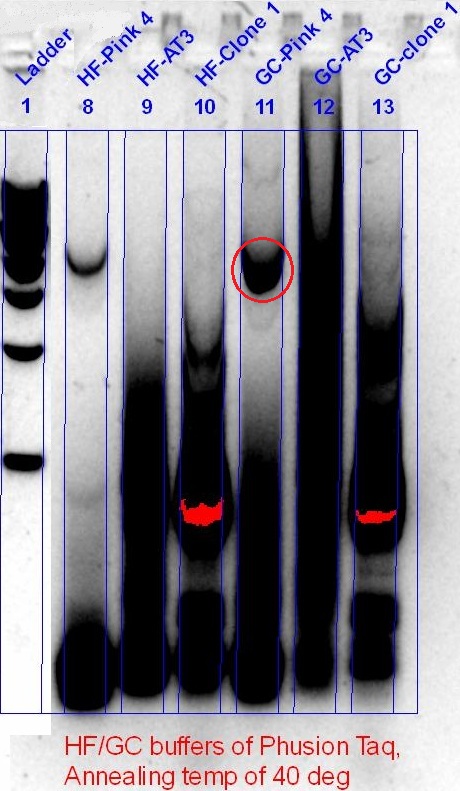
Despite having alkaline phosphatase treatment, and gel purification of the cut plasmid backbone (not shown), the re-ligation of the backbone, and the appearance of the red fluorescent protein, showed that the BioBrick construction of the phytochromes failed. Hence, PCR optimisation to get a better product was performed, this time with a much lower annealing temperature (40 °C), screening a range of different templates that was available.
AT-Bph PCR (left), DR-Bph PCR (right), Red circle - Desired band size
However, we did not have enough time to continue with the rest of the construction as time went out.
In conclusion, we learnt that:
- For PCR, the annealing temperature used should be determined by the annealing region of the primer. In this case, at the start, we have been using annealing temperatures that were too high for the annealing region. As such, only when a lower annealing temperature was used (40 °C) did a good amplification occur.
- A low annealing temperature, however, increases the chances of non-specific amplification, which can be seen in the gel photos. To deal with this, new primers should be designed, where the annealing region to the template is increased, while reducing the upstream region of the restriction cut site.
- For plasmid backbones cut with compatible ends, alkaline phosphatase treated products used should be made fresh each time, to reduce the chances of re-ligation.
Heme Oxygenase 1
The PCR optimisation of the RBS-coupled heme oxygenase 1 coding sequence was successful from the start (see notebook), and much more effort was put into the construction of the HO-BioBrick, and the assembly with a T7 promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_I719005 BBa_I719005] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I712074 BBa_I712074]).
After the initial failure of the ligation of the HO-PCR product with the plasmid backbone, and the lack of specificity of the colony PCR (see notebook), the BioBrick construction pipeline was adopted, using a new batch of alkaline phosphatase treated backbone, and the XbaI restriction digest as a screening method. Despite getting only 1 colony from the transformation, the single colony proved to be right on target, giving us our first successful BioBrick.
(Left) The comparison of the uncut plasmid size (pSC-HO) with the RFP-plasmid (pSC-RFP) and the re-ligated plasmid (pSC-empty) showed a successful insertion of the HO gene; smaller than RFP-plasmid, but larger than the re-ligated plasmid. The XbaI digest gave prove of the gene orientation; a plasmid able to be digested with XbaI.
(Right) Confirmation of the HO-BioBrick ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K646000 BBa_K646000]) was performed, using both PCR and a restriction digest using XbaI and PstI. Although the gel was not well resolved, the similarity of the PCR product size when compared with the original template, and that a gene fragment was cleaved out of the HO-BioBrick, was good enough to conclude that the HO-BioBrick construction was successful.
To test our HO-BioBrick, we assembled it to a T7 promoter, supplied in the iGEM distribution pack (plate 1, location 15N and 6N). Using our alternative assembly method, we digested the T7 promoter bricks with SpeI and PstI, and inserted our HO-BioBrick cut with XbaI and PstI, transformed into Bl21(DE3)Tuner E. coli cells. Selection was done on ampicillin plates (T7 bricks have amp-resistance) that also contained ALA (d-aminolevulinic acid, for heme pathway) and IPTG for induction of protein expression in Bl21(DE3)Tuner E. coli cells. A functional heme oxygenase would be able to metabolise heme into biliverdin, which causes the cells to look green.
PLATE PHOTO NEEDED!!!
 "
"