Team:MIT/Project/
From 2011.igem.org
Project Description
<img src='https://static.igem.org/mediawiki/2011/0/0d/Project_intro.png' style="width:80%">
Current medical practices are only able to scratch the surface of tissue engineering. Fortunately, nature has provided us with robust, cellular systems capable of governing the autonomous formation of complex structures. To pursue control of multicellular systems, we made use of three modules of biological parts. These modules include a signaling module, an internal logic processing module, and an output module for differential adhesion.
For the first module, we engineered the Notch-Delta juxtacrine signaling pathway, a ubiquitous system in animal development. The Notch-Delta system allows for cell-cell communication, a crucial element in symmetry breaking during development. We modified the existing Notch-Delta system to feed into an array of articial, introduced orthogonal molecular circuits, our second module. This module is responsible for integrating these signals to determine a certain cellular state. Once cells have fallen into different states in a desired pattern, one of the outputs from these states would be adhesion in order to hold our cells together to maintain pattern formation. We introduced cadherin, a natural intercellular glue.
To understand and predict multicellular behavior, we developed a simulation framework based on the Synthetic Biology Open Language (SMOL) and CompuCell 3D modelling environment. Our models motivated several circuit designs we subsequently tested in the laboratory. Altogether, our developments establish a paradigm for manipulation of intercellular communication systems to drive self-assembly of ever more complex patterns and tissues.
Delta/Notch Interactions
The Delta/Notch ligand-receptor system is a signaling pathway that is conserved in the overwhelming majority of eukaryotes and in all metazoans, or animals. Both proteins are single-pass transmembrane proteins that can interact with one another to form a heteromeric complex capable of activating transcriptional cascades, specifically in the cell bearing the Notch receptor. Below are a few schematcs of the Delta/Notch system in action.
In the mammalian cells we engineered, there are typically between one and four Delta ligands known as the Delta-Like (DLL) family in addition to several Serrate ligands that are homologous to the Jagged family. While both the Delta-Like and Serrate families are relevant in endogenous systems, the mechanics of Serrate/Notch signaling are typically more variable and dependent on interacting proteins than is the Delta-Like family in mammalian cells. We specifically selected only the Notch-1 receptor and Delta-Like-3 ligand for our project in order to optimize our resources for multiple Delta/Notch experiments. In the future, we intend to probe the usefulness of Serrate engineering and explore how modifier proteins like the Fringe fucosyltransferase enzymes or the Mind Bomb ubiquitin ligase, all of which are capable of modifying multiple aspects of Delta/Notch signaling.
The Delta/Notch signaling system has been implicated to have fundamental roles in many cell fate and developmental decisions, including neuronal differentiation, immune cell differentiation, and binary fate decisions between different germ cell layers. Because of its wide application to tissue differentiation of all varieties and our ability to engineer the intracellular domain of Notch to activate gene transcription from promoters of our choosing, we ultimately selected the Delta/Notch system as our premier cell-cell signaling mechanism.
Device Designs
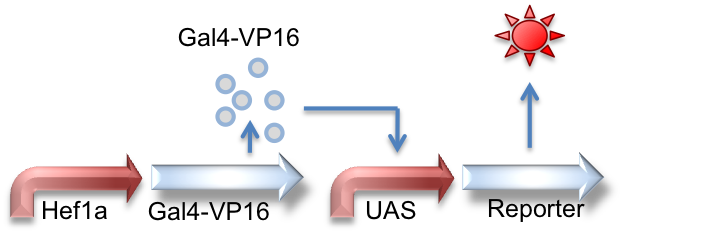
Hef1a:Gal4-VP16, UAS:Reporter
This circuit represents the behavior of the Gal4 activator. Hef1a is a constitutive promoter that will express Gal4-VP16 that can later bind to a UAS promoter, leading to the expression of downstream reporter genes.
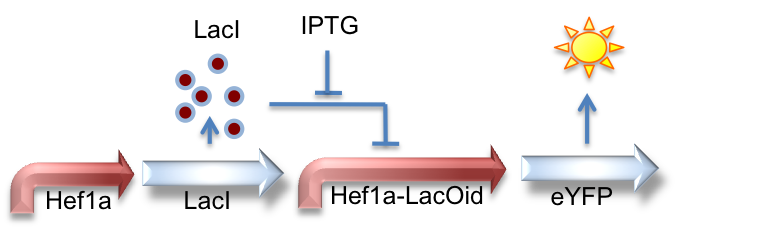
Hef1a:LacI, Hef1a-LacOid:eYFP
Here we represent the activity of the LacI inhibitor. LacI binds to the upstream LacO site and can inhibit expression of downstream reporter genes.
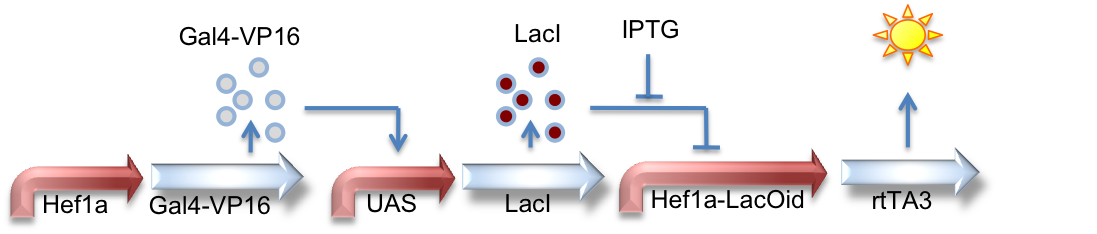
Hef1a:Gal4-VP16, UAS:LacI, Hef1a-LacOid:eYFP
Here we see a circuit that integrates the two systems described above. By tying LacI expression to the Gal4/UAS system, we can activate expression of lacI by GV16 and thus inhibit expression of a reporter gene. This results in overall repression of reporter expression.
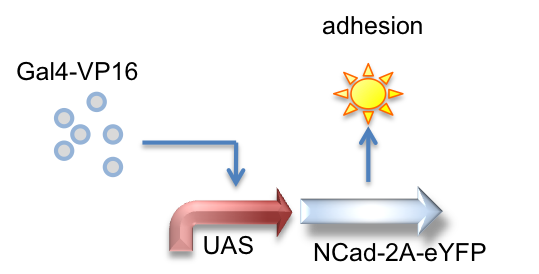
Gal4-VP16, UAS_NCad-2A-eYFP
Here we created a part that is activated by the Gal4-VP16 transactivator to produce N-Cadherin joined to eYFP by a 2A tag, which is cleaved to result in the expression of both proteins.
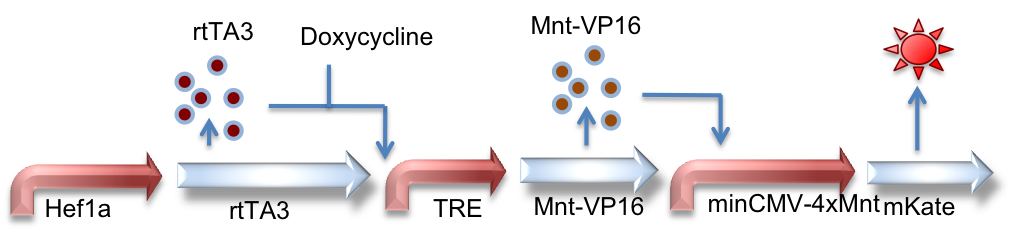
Hef1a:rtTA3, TRE:Mnt-VP16, minCMV-4xMnt:mKate
Here we represent a circuit that has an upstream rtTA3 system feeding into the Mnt activator system. When bound by doxycycline, rtTA3 is capable of inducing expression at the TRE promoter. This in turn leads to expression of the activator Mnt-VP16 which can bind to Mnt sites and activate expression of the reporter gene (mKate, a red fluorescent protein, in this case).
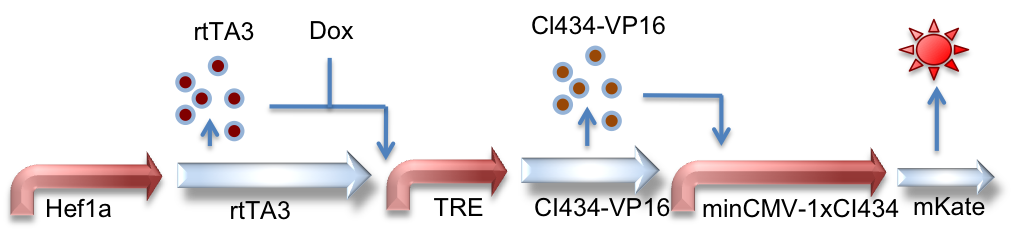
Hef1a:rtTA3, TRE:CI434-VP16, minCMV-1xCI434:mKate
This circuit is very similar in design to the above, instead using a different orthogonal activator-promoter pair: CI434. In this schematic, doxycycline induced expression of CI434 leads to activation of reporter expression downstream of the CI434 binding site (minCMV-1xCI434).
Safety
Would any of your project ideas raise safety issues in terms of:
- researcher safety,
- public safety, or
- environmental safety?
This summer, our team worked on cell patterning in mammalian cells. Part of our team worked with E. Coli in a BL1 lab, and a smaller group worked with mammalian cells in a BL2 lab. Both groups within the team adhered to national and local safety protocols. Extra care was taken not to cross-contaminate lab spaces. Cross contamination from these settings was minimized by designating specific equipment for mammalian cells and for bacteria, as well as immediately changing personal protective equipment when moving between the different lab spaces.
Do any of the new BioBrick parts (or devices) that you made this year raise any safety issues? If yes,
- did you document these issues in the Registry?
- how did you manage to handle the safety issue?
- how could other teams learn from your experience?
Our bacteria and our mammalian cells do not contain any BioBrick parts that code for hazardous proteins or molecules. We also determined that none of our circuits would survive if released outside the lab and thus, the circuits pose no safety concerns to researchers, the public, or the environment.
Is there a local biosafety group, committee, or review board at your institution?
- If yes, what does your local biosafety group think about your project?
The EHS (Environment, Health, and Safety) Office is MIT's biosafety group that enforces lab safety in all labs on campus. They provide safety training, waste management services, and resources for safe lab practices. The safety training included General Biosafety for Researchers, Managing Hazardous Waste, General Chemical Hygiene, lab-specific training, and Bloodborne Pathogen Training. All undergraduates were trained by the EHS to work safely in BL1 labs, and students working with mammalian cells received BL2 lab safety training. EHS is an integral part of our biosafety system and continues to be involved by conducting daily lab safety inspections.
Do you have any other ideas how to deal with safety issues that could be useful for future iGEM competitions? How could parts, devices and systems be made even safer through biosafety engineering?
Our MammoBlock construction standard not only aids in the construction of mammalian parts but also facilitates safer and easier storage of these parts. The MammoBlock standard uses bacterial entry vectors which allows mammalian parts to be stored in BL1 conditions.
Our team is greatly concerned with the safety issues regarding future iGEM research, especially relating to mammalian cells. This standard has allowed us to enter mammalian parts into the registry, and will facilitate the safe shipping and submission of mammalian parts constructed by future iGEM teams. and competitions.
 "
"