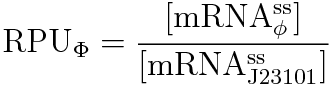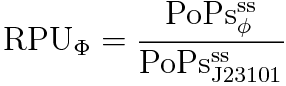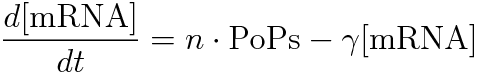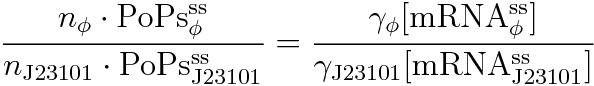Team:Kyoto/Hunger
From 2011.igem.org
(→Method) |
(→Discussion) |
||
| Line 130: | Line 130: | ||
[[file:kyoto_kiga_graph6.png|thumb|left|400px|Fig.6 :graph6]] | [[file:kyoto_kiga_graph6.png|thumb|left|400px|Fig.6 :graph6]] | ||
| + | <div style="width: 100%; float: left;"> | ||
== '''Discussion''' == | == '''Discussion''' == | ||
Graph1 shows the relative amount of RNA expression(GAPDH,TBP,PGK,actin) which is not corrected by internal control gene. | Graph1 shows the relative amount of RNA expression(GAPDH,TBP,PGK,actin) which is not corrected by internal control gene. | ||
| Line 142: | Line 143: | ||
When actin is used as the internal control, the change of GAPDH is little (x0.8~1.0) | When actin is used as the internal control, the change of GAPDH is little (x0.8~1.0) | ||
According from these data, we assumed that GAPDH and actin is suitable for the internal control and that steady state is probably 0~10 minutes. | According from these data, we assumed that GAPDH and actin is suitable for the internal control and that steady state is probably 0~10 minutes. | ||
| + | |||
| + | </div> | ||
== '''Reference''' == | == '''Reference''' == | ||
Revision as of 19:44, 5 October 2011
Contents |
Project Hunger
Introduction
Carnivorous E.coli attracts insects by emitting light, but it is a burden for the E.coli. To reduce this burden, we use nitrogen regulatory proteins, NtrB and NtrC. They activate σ54 promoter when the supply of nitrogen is not enough. NtrB and NtrC are coded in glnL and glnG, respectively.
Ammonia is an essential nitrogen source for the bactria. When enteric bacteria are deprived of ammonia, they express glnA to produce glutamine synthetase(GS) under the σ54promoter. The transcription from σ54promoter is stimilated by phosphorylated form of NtrC(NtrC-P). The σ54 RNA polymerase binds to the glnA promoter, forming a closed complex, but cannot form an open complex and initiate transcription until it is activated by NtrC-P. NtrC is phosphorylated by NtrB-P, an autokinase which phosphorylates itself with ATPs. Phosphorylation and dephosphorylation of NtrB and C are controlled so that cell has sufficient NtrC-P when the concentration of ammoniacal source is low.
The concentration of ammoniacal source is detected by the ratio of αketoglutarate to glutamine. If glutamine levels are low, less αketoglutarate is synthesized by GS and, as a result, Pii retains UMP and so cannot bind to NtrB. NtrB can then phosphorylate itself and transfer this phosphate to NtrC.
[GS Reaction Under Low NH3 Concentration]
Glutamate + NH3 + ADP → Glutamine + ADP + phosphate
GS
Glutamine + αketoglutarate → 2 glutamate
GS
Although this nitrogen controlled switch is complex,
However, the σ54promoter has never been evaluated quantitatively. We characterize the σ54promoter by Relative Promoter Unit (RPU), because absolute promoter activity depends on test conditions and measuring instruments. RPU can reduce this Coefficient of Variation (CV) from 39.1% to 17.5% [2]. Therefore RPU can make it easier for us to share the data of promoter activity and use BioBricks.
We have used GFP fluorescence to measure RPU previously, but this time, we tried to calculate RPU using another way for the following reasons:
- We know that it is a lot of trouble to calculate RPU using GFP fluorescence without platereader.
- Because the σ54promoter relates to the metabolism of glutamine, the concentration of the glutamine changes and activity of σ54promoter changes.
When we calculate RPU using GFP fluorescence, we need to measure GFP fluorescence at two points in an exponential growth phase and same glutamine concentration, but we have the following problems.
- before E.coli reach exponential growth phase, the concentration of glutamine changes
- the concentration of glutamine is different between one point and the other point.
We devised the new way of calculating RPU using the amount of mRNA, according to the following equation.
We can characterize promoters with RPU easily by using this new way.
† NtrB and NtrC are otherwise called NRII, NRI
Method
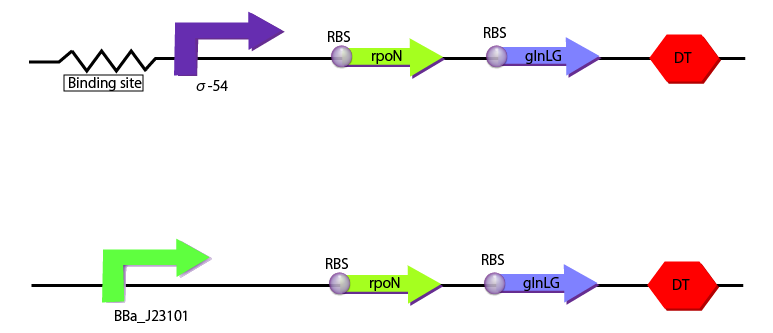
We created the following two constructions to characterize σ54 promoter with RPU depending on the concentration of glutamine. One construction includes BBa_J23101 as a promoter which is used as the standard and the other includes binding sites and σ54 promoter.
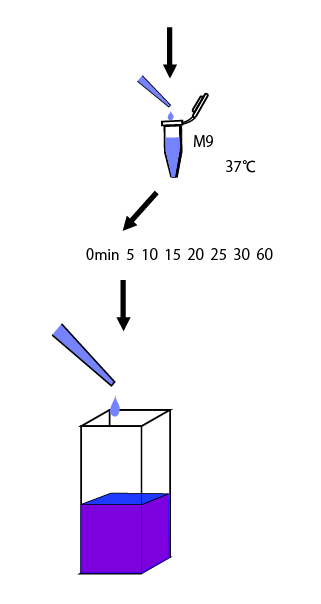
This time we intended to assay the activity of promoter against glutamine concentration. It is desirable to induct after the cultivation in the medium without glutamine. However, from our last year's experience, we knew that E.coli grew really inefficiently in a M9 medium without casamino acids. So, at first we cultivated E.coli in the medium with casamino acids overnight, then changed the medium with the one without casamino acids. After that, we performed an experiment to determine how long it took to reach a steady state.
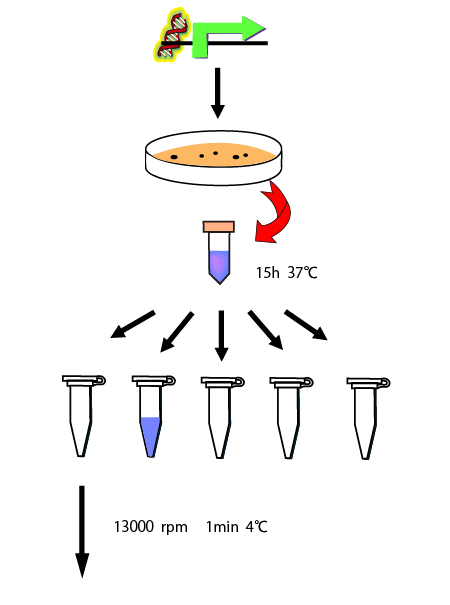
When we use new way to measure RPU, we need to know the length of time to reach steady state. So, we made the following preliminary experiments to measure it.
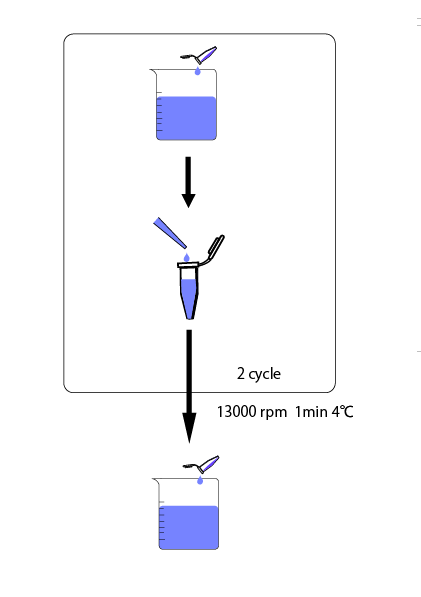
We cultivated E.coli in M9 media(+ casamino acids) for about 15 hours and dispenced 2.4ml to each tube. Then, we centrifuged these tubes (13,000 rpm , 4°C, 1min) and discarded the supernatant. We added 2.4ml media(- casamino acids) and centrifuged at 4°C twice. Again, we centrifuged these tubes and discarded the supernatant and added 2.4ml media(-casamino acids) at 37 °C. We brought up E.coli at 0,5,10,15,20,25,30,60min and extracted RNA and synthesized cDNA. Finally, we used real time PCR.
Result
We get this new equation for measuring RPU.
The derivation is below.
RPU is defined as follows.
PoPS (Polymerase Per Second) is the unit of absolute “promoter activity”. It is defined as the number of RNA polymerase molecules that pass by the final base pair of the promoter and continue along DNA as an elongation complex.
Where:
- [mRNA] is the concentration of mRNA,
- γ is the mRNA degradation rate,
- n is the copy number of the plasmid containing the promoter
Following equations related to promoterφ and J23101 are derived because d[mRNA]/dt = 0.
If the test promoter φ and the reference standard promoter are measured under the same culture conditions and both promoters are carried on the same backbone plasmid, following equations are approved.
Equation(3) divided by (4) is (7) .
We can reduce (7) because of (5) and (6) and use (1). Then we get this equation.
So, we can calculate RPU with this equation.
Discussion
Graph1 shows the relative amount of RNA expression(GAPDH,TBP,PGK,actin) which is not corrected by internal control gene. Although the initial amount of RNA is not equal, it can be said that more than twice the difference is not an accidental error of the experiment. Since the amount of expression of TBP becomes 35 times higher than its initial amount in 15 minutes, we can say that TBP is not suitable for the internal control and that E.coli is not in the steady state after 15 minutes in this experiment.
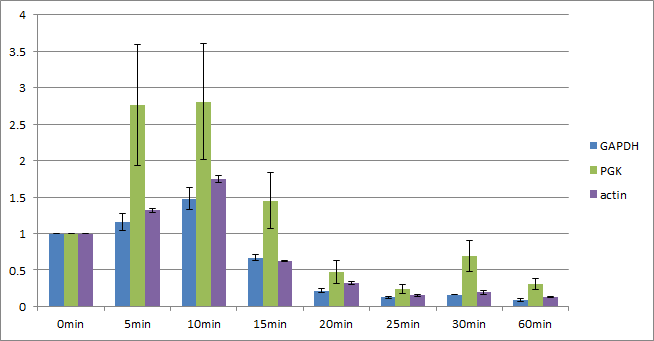
Graph2 is the same data with the graph1 except TBP. Since the expression level of PGK changes a lot, PGK is not suitable for the internal control either. In 0-10 minutes the expression level of GAPDH and actin doesn’t change so much and behave similarly.
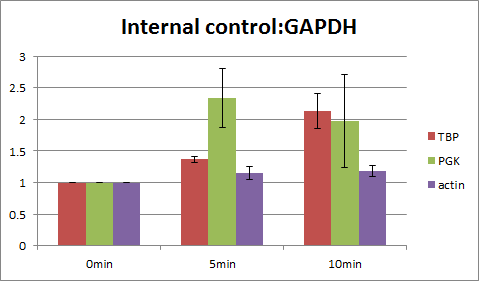
Graph3-6 show the data corrected by each internal control. When GAPDH is used as the internal control, the change of actin is little (x1.0~1.2) When actin is used as the internal control, the change of GAPDH is little (x0.8~1.0) According from these data, we assumed that GAPDH and actin is suitable for the internal control and that steady state is probably 0~10 minutes.
Reference
[1] P. Jiang and A. J. Ninfa, “Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in vitro.,” Biochemistry, vol. 46, no. 45, pp. 12979-96, Nov. 2007.
[2] J. R. Kelly et al., “Measuring the activity of BioBrick promoters using an in vivo reference standard.,” Journal of biological engineering, vol. 3, p. 4, Jan. 2009.
[3] M. Al-Azri, H. Al-Azri, F. Al-Hashmi, S. Al-Rasbi, K. El-Shafie, and A. Al-Maniri, “Factors Affecting the Quality of Diabetic Care in Primary Care Settings in Oman: A qualitative study on patients’ perspectives.,” Sultan Qaboos University medical journal, vol. 11, no. 2. pp. 207-13, May-2011.
[4] L. J. Reitzer and B. Magasanik, “Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter.,” Cell, vol. 45, no. 6, pp. 785-92, Jun. 1986.
[5] B. Magasanik, “Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation.,” Biochimie, vol. 71, no. 9-10, pp. 1005-12, 1989.
[6] J. Keener and S. Kustu, “Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 85, no. 14, pp. 4976-80, Jul. 1988.
[7] J. R. Kelly et al., “Measuring the activity of BioBrick promoters using an in vivo reference standard.,” Journal of biological engineering, vol. 3, p. 4, Jan. 2009.
 "
"