Team:Harvard/Template:NotebookDataJuly3
From 2011.igem.org
(Difference between revisions)
(→Transformation for OZ plasmids) |
|||
| (9 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
'''Thio kan-ZFB-wp-hisura optimizing PCR results:''' | '''Thio kan-ZFB-wp-hisura optimizing PCR results:''' | ||
*The gel stab PCR produced a product of the right size, but the 2kb side product was still brighter. The 2-PCR protocol also still had substantial side products but at least they did not completely overshadow the real product. We will accordingly go ahead and do a gel extraction on the 2-PCR samples so that we can try lambda red with them next week. | *The gel stab PCR produced a product of the right size, but the 2kb side product was still brighter. The 2-PCR protocol also still had substantial side products but at least they did not completely overshadow the real product. We will accordingly go ahead and do a gel extraction on the 2-PCR samples so that we can try lambda red with them next week. | ||
| - | [[File: | + | [[File: HARV2011.07.15.thiokan PCR optimization(labeled).png||thumb|none|Different attempts and thio kan-ZFB-hisura PCR 7/15/11]] |
'''pSR01 growth phenotype results:''' | '''pSR01 growth phenotype results:''' | ||
*Unfortunately the computer hooked up to the plate reader spontaneously restarted during the night to download updates, so we lost all the data. Just by looking at the plate, it still seems like IPTG is inhibiting growth. The ∆HisB∆pyrF∆rpoZ controls also did not grow, probably because the plate the colonies were taken from was too old, so we will restreak from the glycerol stock and set up the plate reader again this weekend. | *Unfortunately the computer hooked up to the plate reader spontaneously restarted during the night to download updates, so we lost all the data. Just by looking at the plate, it still seems like IPTG is inhibiting growth. The ∆HisB∆pyrF∆rpoZ controls also did not grow, probably because the plate the colonies were taken from was too old, so we will restreak from the glycerol stock and set up the plate reader again this weekend. | ||
| Line 13: | Line 13: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.15_electro_miniprep_and_cultures_annotated.png|thumb|Description of gel below.]] |
|} | |} | ||
| Line 39: | Line 39: | ||
{| | {| | ||
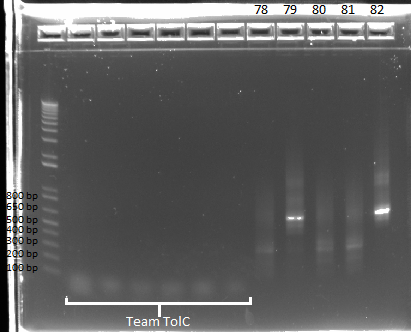
| - | |[[File: | + | |[[File:HARV2011.7.15_Team_TolC(lanes_2-7)_and_omega-ultramer_plasmid_pcr_annotated.png|thumb|Team ZF results in lanes 8-12]] |
|} | |} | ||
| Line 52: | Line 52: | ||
{| | {| | ||
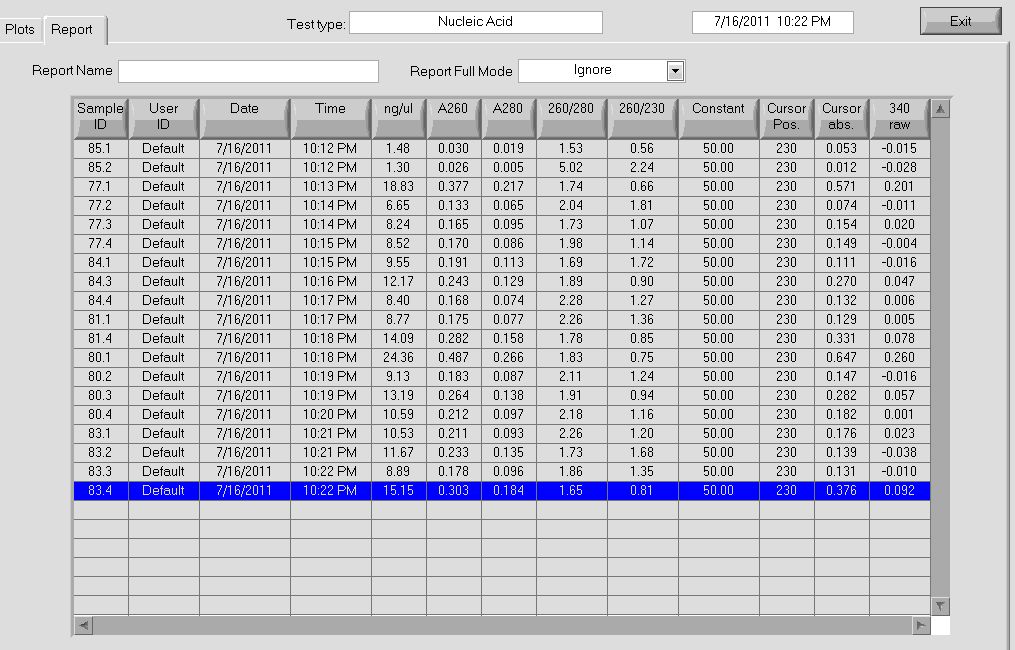
| - | |[[File: | + | |[[File:HARV2011.7.16_Colony_Miniprep_Conc.jpg|thumb|Plasmid concentrations]] |
|} | |} | ||
====PCR on minipreps to check cross junction==== | ====PCR on minipreps to check cross junction==== | ||
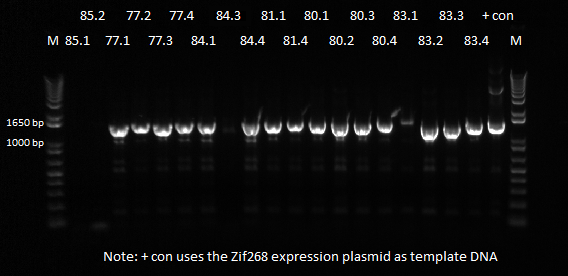
| - | PCR was done on the miniprep products to check for the cross junction length, expected band size of ~1.4 kb. The protocol used was the same as | + | PCR was done on the miniprep products to check for the cross junction length, expected band size of ~1.4 kb. The protocol used was the same as the previous time. In addition to PCR-ing the miniprep products, a positive control was also included, which was the finished Zif268 + omega + spec backbone plasmid (this was the first successful junction PCR we obtained, and so it should work again). |
====Replating final samples for 78, 79, 82==== | ====Replating final samples for 78, 79, 82==== | ||
| Line 69: | Line 69: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.17_miniprep_junction_pcr_darker_annotated.png|thumb|PCR Results, expected band at ~1.4 kb]] |
|} | |} | ||
| Line 82: | Line 82: | ||
{| | {| | ||
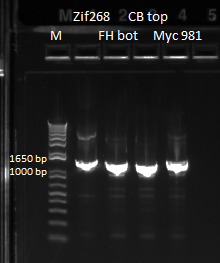
| - | |[[File: | + | |[[File:HARV2011.7.18_junction_primer_pcr_for_seq_annotated.png|thumb|Gel of cross junction of Zif268, FH bottom, CB top, Myc 981]] |
|} | |} | ||
| Line 112: | Line 112: | ||
===Team Wolfe=== | ===Team Wolfe=== | ||
'''SNOWMAN!''' | '''SNOWMAN!''' | ||
| - | [[File: | + | [[File: HARVSnowman1.jpeg|left|Our snowman Persikov]] |
| - | [[File: | + | [[File: HARVSnowman3.jpeg|none|Brandon with his creation]] |
'''EcNR2 lambda red (Noah's prep):''' | '''EcNR2 lambda red (Noah's prep):''' | ||
| Line 130: | Line 130: | ||
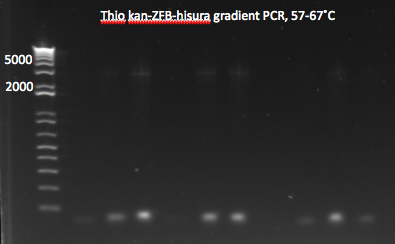
**same as above but with 12 reactions, annealing temperatures from 57-67˚C | **same as above but with 12 reactions, annealing temperatures from 57-67˚C | ||
*Results: most of the gradient reactions produced a faint but clean band at 3kb. The touch down PCR had a stronger 3kb band but also lots of side products. | *Results: most of the gradient reactions produced a faint but clean band at 3kb. The touch down PCR had a stronger 3kb band but also lots of side products. | ||
| - | [[File: | + | [[File: HARV2011.07.18.thiokan grad1(labeled).png|thumb|left|Thio kan-ZFB-hisura (Noah's) gradient part 1 7/18/11]] |
| - | [[File: | + | [[File: HARV2011.07.18.thiokan grad2+td(labeled).png|none|Thio kan-ZFB-hisura (Noah's) gradient part 2 and touchdown 7/18/11]] |
===Team TolC=== | ===Team TolC=== | ||
| Line 148: | Line 148: | ||
**Placed colonies in 20 μL of water and used 1 μl for a 20 μL PCR reaction | **Placed colonies in 20 μL of water and used 1 μl for a 20 μL PCR reaction | ||
**Then took 10 μL from 20 μL dilution and added to 100 μL of LB in 96 well plate and put in 30°C fridge, at 3:19 pm | **Then took 10 μL from 20 μL dilution and added to 100 μL of LB in 96 well plate and put in 30°C fridge, at 3:19 pm | ||
| - | **Same protocol as [ | + | **Same protocol as [https://2011.igem.org/Team:Harvard/Protocols#PCR KAPA PCR] with the following changes made |
***Step 3, gradient for annealing, 55°C - 77°C, for a range of 55,57,59,61 and 63°C for the two rows of 5 PCR tubes each (placed leftmost) | ***Step 3, gradient for annealing, 55°C - 77°C, for a range of 55,57,59,61 and 63°C for the two rows of 5 PCR tubes each (placed leftmost) | ||
***Step 4, Extension, 1 min | ***Step 4, Extension, 1 min | ||
| Line 154: | Line 154: | ||
**Name: TOLCGRAD, PCR5 machine.</div> | **Name: TOLCGRAD, PCR5 machine.</div> | ||
<div id="719" style="display:none"> | <div id="719" style="display:none"> | ||
| + | |||
==July 19th== | ==July 19th== | ||
===Team ZF=== | ===Team ZF=== | ||
| Line 162: | Line 163: | ||
====Minipreps of colonies and controls==== | ====Minipreps of colonies and controls==== | ||
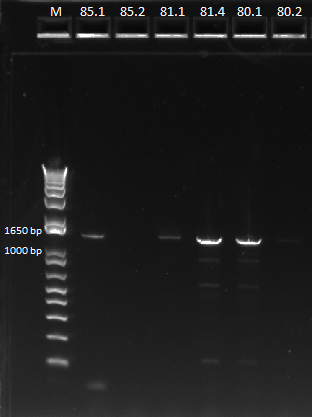
| - | Our minipreps from | + | Our minipreps from Saturday had low yields, especially for culture 85. We suspect this is why the PCR [[#Gel analysis of miniprep junction PCR on Sunday did not work for this sample. Unfortunately, 85.3 and 85.4 did not grow even after being left in the shaker overnight. These were two cultures we made by picking new colonies from plate 85. This may have occurred because we did not actually pick the colony, or the colony was a cheater. Thus, we performed minipreps on 80.1, 80.2, 81.1, 81.4 (the controls, chosen for their wide range of concentrations after our first miniprep), and 85.1 and 85.2. |
In order to figure our why our minipreps were resulting in unexpectedly low yields and increase our yields: | In order to figure our why our minipreps were resulting in unexpectedly low yields and increase our yields: | ||
| Line 175: | Line 176: | ||
{| | {| | ||
| - | |[[File: | + | |[[File: HARV2011.7.18_Miniprep_redo.jpg |thumb|Concentration and purity of our miniprep product]] |
|} | |} | ||
====PCR of miniprep products==== | ====PCR of miniprep products==== | ||
| - | We then ran a PCR for the cross-junction on all 6 (80.1, 80.2, 81.1, 81.4, 85.1, 85.2) samples, using | + | We then ran a PCR for the cross-junction on all 6 (80.1, 80.2, 81.1, 81.4, 85.1, 85.2) samples, using our usual cross junction protocol. |
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.19_miniprep_test_annotated.png|thumb|Gel results, expected product size of 1.4 kb]] |
|} | |} | ||
| Line 191: | Line 192: | ||
====Designing ultramers for Zif268, OZ052, and OZ123==== | ====Designing ultramers for Zif268, OZ052, and OZ123==== | ||
| - | In preparation for designing our 96-well practice plate, we designed ultramers for our three positive controls Zif268, OZ052, and OZ123. These ultramers contain the Type II binding sites as well as the F2/F3 fingers. Since the | + | In preparation for designing our 96-well practice plate, we designed ultramers for our three positive controls Zif268, OZ052, and OZ123. These ultramers contain the Type II binding sites as well as the F2/F3 fingers. Since the practice plate will contain oligos that encode for F1, we can use these three positive controls to practice and ensure that we secure the technique of using the Type II binding sites to swap in F1 into our expression plasmids. The ultramers have not yet been ordered. |
===Team TolC=== | ===Team TolC=== | ||
| Line 203: | Line 204: | ||
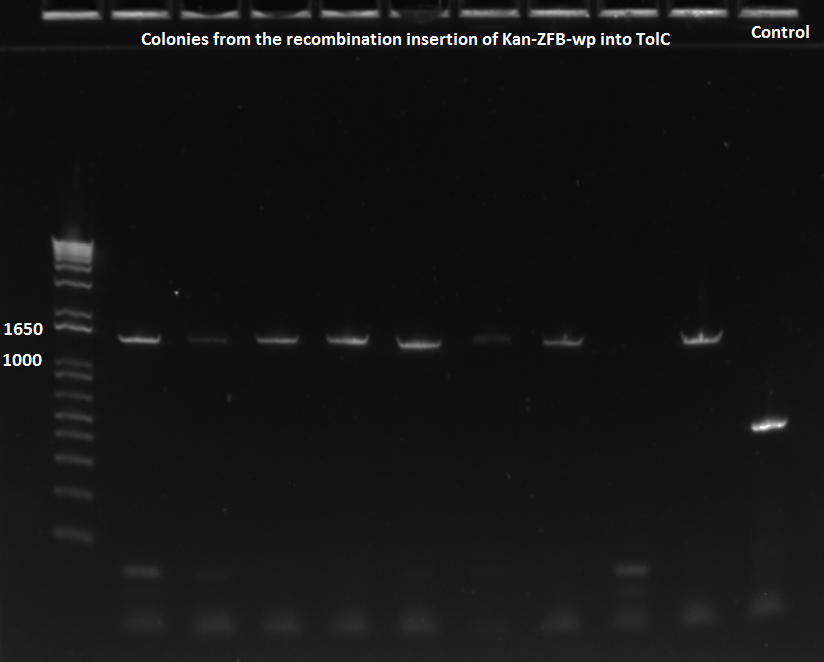
*5 of ZF090, 4 of ZF091, and 1 from old plate were run in a PCR with 62˚C annealing temperature | *5 of ZF090, 4 of ZF091, and 1 from old plate were run in a PCR with 62˚C annealing temperature | ||
*primers used were TolC_seq_F and TolC_seq_R | *primers used were TolC_seq_F and TolC_seq_R | ||
| - | [[File: | + | [[File: HARV2011.07.19.Noahkan-ZFBinsertionsuccessPCR.png|thumb|none|EcNR2 lambda red 1529620 PCR 7/19/11]] |
| - | [[File: | + | [[File: HARV2011.07.19.Noahkan-ZFBinsertionsuccessPCR2.png|thumb|none|EcNR2 lambda red 1529620 PCR 7/19/11]] |
*Chose colonies based on the intensity of the bands and grew up cultures of ZF091-2d, ZF091-4d, ZF090-1d, ZF091-5d, ZF090-1s, ZF090-2s, ZF090-3s, and ZF091-3s | *Chose colonies based on the intensity of the bands and grew up cultures of ZF091-2d, ZF091-4d, ZF090-1d, ZF091-5d, ZF090-1s, ZF090-2s, ZF090-3s, and ZF091-3s | ||
| Line 223: | Line 224: | ||
*PCR showed that colonies 1-3 had the insert while 4 did not. Yay! (Note that in the gel image, we accidentally cut off the 300bp band in lane 4) | *PCR showed that colonies 1-3 had the insert while 4 did not. Yay! (Note that in the gel image, we accidentally cut off the 300bp band in lane 4) | ||
*We will grow up a reinoculated culture of colony 1 for a glycerol stock. If the Wolfe strain keeps not working, we at least have the insert in an EcNR2 line and can hopefully MAGE out the other genes without too much difficulty. | *We will grow up a reinoculated culture of colony 1 for a glycerol stock. If the Wolfe strain keeps not working, we at least have the insert in an EcNR2 line and can hopefully MAGE out the other genes without too much difficulty. | ||
| - | [[File: | + | [[File: HARV2011.07.19.EcNR2 lambda red insert(labeled).png|thumb|none|EcNR2 lambda red 1529620 PCR 7/19/11]] |
'''pSR01 growth phenotype:''' | '''pSR01 growth phenotype:''' | ||
*We will try to used the plate reader again to compare the growth phenotypes of ∆HisB∆pyrF∆rpoZ and ∆HisB∆pyrF∆rpoZ+pSR01. Used the same media as last time but since we're using a 48 well plate, each well has 250µL media. We used 5 pSR01 colonies and one selection strain colony as a control. | *We will try to used the plate reader again to compare the growth phenotypes of ∆HisB∆pyrF∆rpoZ and ∆HisB∆pyrF∆rpoZ+pSR01. Used the same media as last time but since we're using a 48 well plate, each well has 250µL media. We used 5 pSR01 colonies and one selection strain colony as a control. | ||
| Line 308: | Line 309: | ||
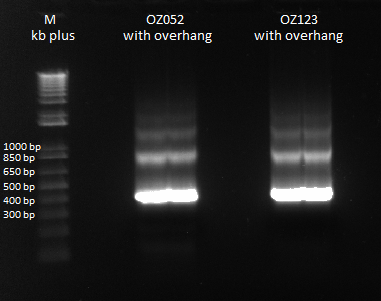
====Construction of OZ052 and OZ123: Joining the Finger with Homology regions==== | ====Construction of OZ052 and OZ123: Joining the Finger with Homology regions==== | ||
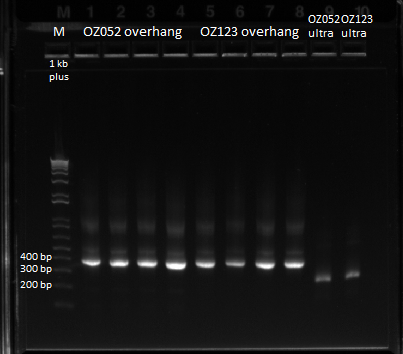
| - | Previously, we did a PCR to | + | Previously, we did a PCR to reassemble the ultramers for OZ052 and OZ123, but they only coded strictly for F1/F2/F3 and did not have any overhang homology with any surrounding code. Thus, we performed PCR to add these homology overhangs so that we can integrate them with the omega subunit as well as the spec backbone to create a final expression plasmid. |
The program we used was 55* annealing temperature, and a 15 second extension time. Gel results pictured below. | The program we used was 55* annealing temperature, and a 15 second extension time. Gel results pictured below. | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.20_OZ_overhang_additions_annotated.png|thumb|Overlap PCR to extend ultramers with homologies]] |
|} | |} | ||
| Line 360: | Line 361: | ||
All of the cultures grew overnight, so we performed a miniprep on them to obtain the plasmids. | All of the cultures grew overnight, so we performed a miniprep on them to obtain the plasmids. | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.21_Bad_sequence_redo_miniprep.jpg|thumb|Plasmid concentrations after miniprep]] |
|} | |} | ||
Our concentrations were better than they have been in the past; notably, after the addition of buffer P2, our solution actually turned a bright blue (indicating lysis of the cells), which we had not seen before. | Our concentrations were better than they have been in the past; notably, after the addition of buffer P2, our solution actually turned a bright blue (indicating lysis of the cells), which we had not seen before. | ||
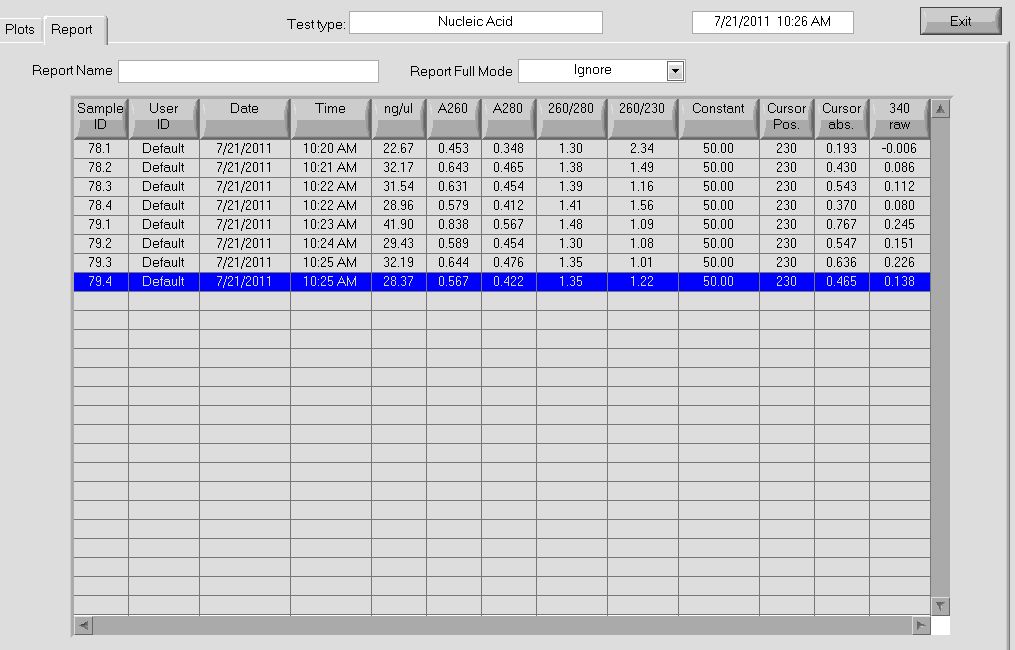
| - | We then ran a PCR on our miniprep product, to check for our desired insert. We used our | + | We then ran a PCR on our miniprep product, to check for our desired insert. We used our usual protocol for a PCR of the expression plasmid cross-junction. |
{| | {| | ||
| - | |[[File: | + | |[[File: HARV2011.7.21_New_colonies_junction_pcr_annotated.png|thumb|PCR results, note that some lanes do not show strong bands]] |
|} | |} | ||
| Line 374: | Line 375: | ||
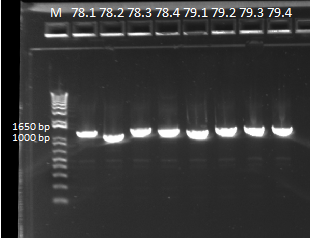
====Sending out our expression colonies for sequencing==== | ====Sending out our expression colonies for sequencing==== | ||
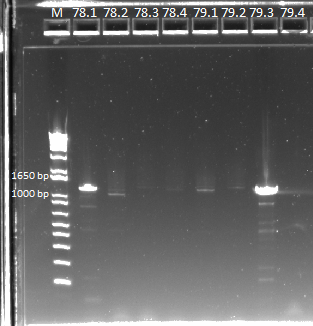
| - | We sent out the samples from | + | We sent out the samples from yesterday that had no more than 1 SNP for reverse sequencing. In addition, we also sent out 78 and 79 for forward and reverse sequencing. |
*Reverse sequencing: 77.1, 77.3, 77.4, 78.1, 80.1, 80.2, 80.4, 81.1, 81.4, 82.1, 83.4, 84.1, 84.4, Zif268 | *Reverse sequencing: 77.1, 77.3, 77.4, 78.1, 80.1, 80.2, 80.4, 81.1, 81.4, 82.1, 83.4, 84.1, 84.4, Zif268 | ||
*Forward and reverse sequencing: 781, 78.2, 78.3, 78.4, 79.1, 79.2, 79.3, 79.4 | *Forward and reverse sequencing: 781, 78.2, 78.3, 78.4, 79.1, 79.2, 79.3, 79.4 | ||
| Line 383: | Line 384: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.21_more_sequencing_pcr_ver_2_annotated.png|thumb|PCR results, looking good]] |
|} | |} | ||
| Line 389: | Line 390: | ||
====Gel Extraction and Isothermal Assembly of OZ fingers+homology==== | ====Gel Extraction and Isothermal Assembly of OZ fingers+homology==== | ||
| - | We ran a gel extraction of our PCR product from | + | We ran a gel extraction of our PCR product from yesterday, so that we could use it for isothermal assembly later. Strangely, we got bands that were larger than our largest theoretical product. |
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.21_OZ_overhang_gel_extraction_annotated.png|thumb|Prepare yourself to be gel-extracted]] |
|} | |} | ||
| Line 422: | Line 423: | ||
*Also replated the 7/20 lambda red culture to see if we can get any good cells (1.5mL).</div> | *Also replated the 7/20 lambda red culture to see if we can get any good cells (1.5mL).</div> | ||
<div id="722" style="display:none"> | <div id="722" style="display:none"> | ||
| + | |||
==July 22nd== | ==July 22nd== | ||
===Team ZF=== | ===Team ZF=== | ||
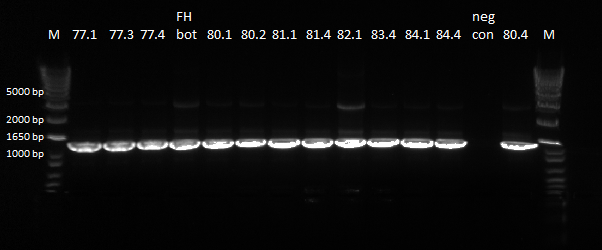
| Line 452: | Line 454: | ||
| - | To this end, we redid the PCRs for everything in preparation for the resequencing, using the junction primers and | + | To this end, we redid the PCRs for everything in preparation for the resequencing, using the junction primers and the previous protocol. The resulting gels can be observed below: |
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.22_78_and_79_for_sequencing_annotated.png|thumb|E-gel showing bands for samples 78.1-78.4 and 79.1-79.4, expected size of 1.4 kb]] |
|} | |} | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.22_rest_of_pcr_for_sequencing_annotated.png|thumb|Remaining samples, expected size of 1.4 kb]] |
|} | |} | ||
| Line 467: | Line 469: | ||
====Transformation for OZ plasmids==== | ====Transformation for OZ plasmids==== | ||
| - | Today, we took the results of the isothermal assembly from yesterday and did chemical transformation to introduce the plasmids into the Top 10 ChemComp bacteria, using [ | + | Today, we took the results of the isothermal assembly from yesterday and did chemical transformation to introduce the plasmids into the Top 10 ChemComp bacteria, using [https://2011.igem.org/Team:Harvard/Protocols#Cultures our protocol for chemical transformations]. They were left to recover for approximately 1.5 hours. We plated 2 dilutions, 10 ul and 50 ul, and left them overnight at 37* in the incubator. |
===Team Wolfe=== | ===Team Wolfe=== | ||
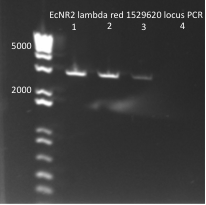
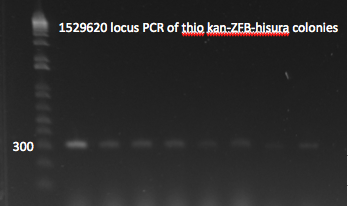
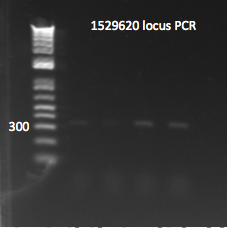
*Yesterday's lambda red plate had several large colonies--but so did the negative control plate. We will go ahead and do a PCR of the 1529620 locus (as previously described) on 12 of the colonies in case one of them happens to have the insert. The colonies were also grown up simultaneously in LB+kan. | *Yesterday's lambda red plate had several large colonies--but so did the negative control plate. We will go ahead and do a PCR of the 1529620 locus (as previously described) on 12 of the colonies in case one of them happens to have the insert. The colonies were also grown up simultaneously in LB+kan. | ||
**Results: all of the colonies were cheaters | **Results: all of the colonies were cheaters | ||
| - | [[File: | + | [[File: HARV2011.07.22.thiokan insert check1(labeled).png|thumb|left|1529620 locus PCR 7/22/11]] |
| - | [[File: | + | [[File: HARV2011.07.22.thiokan insert check2(labeled).png|thumb|none|1529620 locus PCR part 2 7/22/11]] |
*In case these colonies do not have the insert, as we suspect, we will repeat lambda red with the thio kan-ZFB-wp-his3-ura3 insert | *In case these colonies do not have the insert, as we suspect, we will repeat lambda red with the thio kan-ZFB-wp-his3-ura3 insert | ||
**1mL culture for each condition, 210 ng DNA | **1mL culture for each condition, 210 ng DNA | ||
| Line 481: | Line 483: | ||
====Gel for Jun's MAGE rpoZ, rowD==== | ====Gel for Jun's MAGE rpoZ, rowD==== | ||
*The gel appeared to have bands, so we are running a second PCR to confirm it</div> | *The gel appeared to have bands, so we are running a second PCR to confirm it</div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
 "
"