|
|
| Line 1: |
Line 1: |
| - | <div style="display:none"> | + | <div id='810' style="display:none"> |
| | ==August 10== | | ==August 10== |
| | ===Team TolC=== | | ===Team TolC=== |
| Line 11: |
Line 11: |
| | ***numbered, from 1-6, 1-3 is colony 1, 4-6 is colony 2. 1,4=TT 2,5=RR, 3,6=ZR | | ***numbered, from 1-6, 1-3 is colony 1, 4-6 is colony 2. 1,4=TT 2,5=RR, 3,6=ZR |
| | ***PCR successful!! We have the real WN strain now. It should be ready for transformation tomorrow with the zif268 spec plasmid | | ***PCR successful!! We have the real WN strain now. It should be ready for transformation tomorrow with the zif268 spec plasmid |
| - | [[File: 2011.08.10.WN1tt2rr3zrworks(labeled).png|thumb|none|Zeocin/rpoZ and TolC PCR of lambda red colonies 8/10/11]] | + | [[File: HARV2011.08.10.WN1tt2rr3zrworks(labeled).png|thumb|none|Zeocin/rpoZ and TolC PCR of lambda red colonies 8/10/11]] |
| | | | |
| | ===Team Wolfe=== | | ===Team Wolfe=== |
| Line 25: |
Line 25: |
| | '''Results''' | | '''Results''' |
| | *rpoZ was knocked out! | | *rpoZ was knocked out! |
| - | [[File: 2011.08.10.rpoz KO check(labeled).png|thumb|none|Zeocin/rpoZ PCR of lambda red colonies 8/10/11]] | + | [[File: HARV2011.08.10.rpoz KO check(labeled).png|thumb|none|Zeocin/rpoZ PCR of lambda red colonies 8/10/11]] |
| | *Reinoculated LB+zeo with culture 9 to use tomorrow for glycerol stocks and transformations | | *Reinoculated LB+zeo with culture 9 to use tomorrow for glycerol stocks and transformations |
| - |
| |
| - |
| |
| - |
| |
| | | | |
| | ====Digestion of pSB4K5 Plasmid==== | | ====Digestion of pSB4K5 Plasmid==== |
| | *Gel with yesterday's digestion product suggests that neither of the two restriction enzymes (Xba1 or Spe1) cut our plasmid. This was determined by comparing the band sizes to those obtained by midiporep of the pSB4K5 (whole) plasmid. | | *Gel with yesterday's digestion product suggests that neither of the two restriction enzymes (Xba1 or Spe1) cut our plasmid. This was determined by comparing the band sizes to those obtained by midiporep of the pSB4K5 (whole) plasmid. |
| | *Plan of action: We will use smaller reactions to increase the relative concentration of enzymes and we will use slightly more BSA buffer. | | *Plan of action: We will use smaller reactions to increase the relative concentration of enzymes and we will use slightly more BSA buffer. |
| - | [[File:2011.08.10.pSB4K5 digest(labeled).png|thumb|none|pSB4K5 digestion with Spe1 and Xba1]] | + | [[File:HARV2011.08.10.pSB4K5 digest(labeled).png|thumb|none|pSB4K5 digestion with Spe1 and Xba1]] |
| | | | |
| | '''Today's Digestion of pSB4K5 Plasmid''' | | '''Today's Digestion of pSB4K5 Plasmid''' |
| Line 54: |
Line 51: |
| | **1.5µL Spe1 | | **1.5µL Spe1 |
| | *Results: the size of the bands isn't what we expected (backbone should be ~3.4kb, insert ~1.2), but the comparison between the different digestions and the uncut plasmid does imply that the digestion we worked. We will go ahead and gel extract from the Spe+Xba digest and use both ligation and sequencing to be sure that this is indeed the plasmid we want. | | *Results: the size of the bands isn't what we expected (backbone should be ~3.4kb, insert ~1.2), but the comparison between the different digestions and the uncut plasmid does imply that the digestion we worked. We will go ahead and gel extract from the Spe+Xba digest and use both ligation and sequencing to be sure that this is indeed the plasmid we want. |
| - | [[File:2011.08.10.psb4k5 spexba digest(labeled).png|thumb|none|pSB4K5 digest with Spe1, Xba1, and both 8/10/11]] | + | [[File:HARV2011.08.10.psb4k5 spexba digest(labeled).png|thumb|none|pSB4K5 digest with Spe1, Xba1, and both 8/10/11]] |
| | | | |
| | ===Team ZF=== | | ===Team ZF=== |
| | ====Mini library ligation results==== | | ====Mini library ligation results==== |
| - | We checked our plates from our practice plate library assembly [[Lab_Notebook:_August#Practice_plate_oligo_pool|yesterday]], and we got colonies!! Strangely, our 450 ul plate didn't have a huge number more colonies than the 50 ul plate. Dan suggested that this was because we plated all the cells in a 450 ul volume, which may have sloshed around and taken cells with it. Next time we should pellet the cells and resuspend in 100 ul and plate that. Based on our colony numbers with 50 ul, we should probably plate with 100 ul in the future. | + | We checked our plates from our practice plate library assembly yesterday, and we got colonies!! Strangely, our 450 ul plate didn't have a huge number more colonies than the 50 ul plate. Dan suggested that this was because we plated all the cells in a 450 ul volume, which may have sloshed around and taken cells with it. Next time we should pellet the cells and resuspend in 100 ul and plate that. Based on our colony numbers with 50 ul, we should probably plate with 100 ul in the future. |
| | | | |
| | Due to the proliferation of colonies, we picked 90 colonies with the assistance of Will. These 90 colonies should hopefully represent all 18 oligos in equal amounts. We did colony PCRs on all 90 with the junction PCR protocol, and sent them out for sequencing today. We saved these 90 colonies by adding LB/spec to each of the wells after suspending them in the 10 ul of water for the colony PCR. We will eventually harvest and gather all 18 assembled plasmids from this step. | | Due to the proliferation of colonies, we picked 90 colonies with the assistance of Will. These 90 colonies should hopefully represent all 18 oligos in equal amounts. We did colony PCRs on all 90 with the junction PCR protocol, and sent them out for sequencing today. We saved these 90 colonies by adding LB/spec to each of the wells after suspending them in the 10 ul of water for the colony PCR. We will eventually harvest and gather all 18 assembled plasmids from this step. |
| Line 114: |
Line 111: |
| | | | |
| | ====Library ligation optimization==== | | ====Library ligation optimization==== |
| - | Dan suggests that we optimize the vector:insert ratio for our library assembly. We will try using the following insert ratios (assuming a vector unit of 1): 1, 2, 3, 4, 8, 10, 16. Unfortunately, today we did not have enough cut CB bottom plasmid to immediately begin the ligation and transformation, so we attempted to cut more CB bottom. After performing a cut with Bsa-HF using the [[Lab_Notebook:_August#New_restriction_enzyme_protocol|previous protocol]], we performed a PCR purification on the product, only to find that there were only 3.4 ng/ul of cut plasmid present. This is not enough to perform the ligation, and so we will have to try the CB bottom plasmid cut again tomorrow morning when the cultures have grown enough. We can then do a ligation and transformation tomorrow and count colonies on Friday. | + | Dan suggests that we optimize the vector:insert ratio for our library assembly. We will try using the following insert ratios (assuming a vector unit of 1): 1, 2, 3, 4, 8, 10, 16. Unfortunately, today we did not have enough cut CB bottom plasmid to immediately begin the ligation and transformation, so we attempted to cut more CB bottom. After performing a cut with Bsa-HF using the previous protocol, we performed a PCR purification on the product, only to find that there were only 3.4 ng/ul of cut plasmid present. This is not enough to perform the ligation, and so we will have to try the CB bottom plasmid cut again tomorrow morning when the cultures have grown enough. We can then do a ligation and transformation tomorrow and count colonies on Friday. |
| | | | |
| - | As stated in the earlier section, we should probably plate 100 ul of transformed bacteria to standardize everything and to keep colony counting fairly manageable. | + | As stated in the earlier section, we should probably plate 100 ul of transformed bacteria to standardize everything and to keep colony counting fairly manageable.</div> |
| | + | <div id='811' style="display:none"> |
| | ==August 11th== | | ==August 11th== |
| | ===Team Wolfe=== | | ===Team Wolfe=== |
| Line 203: |
Line 201: |
| | *Since colonies are not near mid-log we may have to fill plate reader tomorrow and run then as well | | *Since colonies are not near mid-log we may have to fill plate reader tomorrow and run then as well |
| | ====Gradient Plates==== | | ====Gradient Plates==== |
| - | *Created 5 50 mL total gradient plates with 2x Kan in the wedge layer, 20ul of Bluejuice in each 25ml of LB+agar+Kan to mark it with a dye. | + | *Created 5 50 mL total gradient plates with 2x Kan in the wedge layer, 20ul of Bluejuice in each 25ml of LB+agar+Kan to mark it with a dye.</div> |
| - | | + | <div id='812' style="display:none"> |
| | ==August 12th== | | ==August 12th== |
| | ===Team Wolfe=== | | ===Team Wolfe=== |
| Line 217: |
Line 215: |
| | **Team ZF's "CJUN" protocol: pZE23G 2123R and pZE23G 3581F primers | | **Team ZF's "CJUN" protocol: pZE23G 2123R and pZE23G 3581F primers |
| | **Note well 5 is fainter but is probably due to a loading error where a small volume of sample was left out. | | **Note well 5 is fainter but is probably due to a loading error where a small volume of sample was left out. |
| - | [[File: 2011.08.12.cjun(labeled).png|thumb|none|EcNR2 selection strain colonies transformed with zif268 8/12/11]] | + | [[File: HARV2011.08.12.cjun(labeled).png|thumb|none|EcNR2 selection strain colonies transformed with zif268 8/12/11]] |
| | | | |
| | ====Selection strain genotype confirmation==== | | ====Selection strain genotype confirmation==== |
| Line 261: |
Line 259: |
| | | | |
| | ====Isothermal assembly and transformation of GFP reporter==== | | ====Isothermal assembly and transformation of GFP reporter==== |
| - | We ran our PCR product from [[#PCR to get out GFP| yesterday]] on a 1% agarose gel. We got an appropriate band at approximately 700 bp. (Unfortunately, we could not get the imaging software to work at the time, so no image is available) | + | We ran our PCR product from yesterday on a 1% agarose gel. We got an appropriate band at approximately 700 bp. (Unfortunately, we could not get the imaging software to work at the time, so no image is available) |
| | | | |
| | We performed a gel extraction on our GFP PCR product, with the following yields: | | We performed a gel extraction on our GFP PCR product, with the following yields: |
| Line 276: |
Line 274: |
| | | | |
| | ====CB Bottom Digestion==== | | ====CB Bottom Digestion==== |
| - | We digested our CB Bottom plasmid, using our usual recipe. We ran the digestion for 2 hours at 37*, 20 minutes at 65*, and then let the reaction sit at 4* overnight. | + | We digested our CB Bottom plasmid, using our usual recipe. We ran the digestion for 2 hours at 37*, 20 minutes at 65*, and then let the reaction sit at 4* overnight.</div> |
| - | | + | <div id='813' style="display:none"> |
| - | ====Western Blot results==== | + | |
| - | | + | |
| | ==August 13th== | | ==August 13th== |
| | ===Team ZF=== | | ===Team ZF=== |
| Line 339: |
Line 335: |
| | |} | | |} |
| | | | |
| - | Combined with the sequencing results from [[#Check oligo pool sequencing results|before]], we are missing oligos 3, 13, 14, and 16. | + | Combined with the sequencing results from before, we are missing oligos 3, 13, 14, and 16. |
| | | | |
| | ====Results of GFP transformation==== | | ====Results of GFP transformation==== |
| Line 345: |
Line 341: |
| | | | |
| | ====PCR purification of CB Bottom digestion product==== | | ====PCR purification of CB Bottom digestion product==== |
| - | We performed a PCR purification on the CB Bottom digestion, with a yield of 68.96 ng/ul and a 260/280 of 1.85. This will be used in a ligation in the future. | + | We performed a PCR purification on the CB Bottom digestion, with a yield of 68.96 ng/ul and a 260/280 of 1.85. This will be used in a ligation in the future.</div> |
| - | | + | <div id='815' style="display:none"> |
| | ==August 15== | | ==August 15== |
| | ===TolC=== | | ===TolC=== |
| Line 354: |
Line 350: |
| | Yay!!! | | Yay!!! |
| | | | |
| - | [[File: 3AT.png|thumb|none|Selection strain with and without plasmid, and 3-AT 8/12/11]] | + | [[File: HARV3AT.png|thumb|none|Selection strain with and without plasmid, and 3-AT 8/12/11]] |
| | | | |
| | Interestingly, our 5-FOA concentrations did not seem very potent: 3-AT was more effective at retarding growth. We might drop the URA3 part of our selection and instead use higher amounts of 3-AT to distinguish between true hits and false positives. | | Interestingly, our 5-FOA concentrations did not seem very potent: 3-AT was more effective at retarding growth. We might drop the URA3 part of our selection and instead use higher amounts of 3-AT to distinguish between true hits and false positives. |
| | | | |
| - | [[File: 2011.08.12.EcNR2 3AT+5FOA.png|thumb|left|Selection strain without Zif268 with 3-AT and 5-FOA 8/12/11]] | + | [[File: HARV2011.08.12.EcNR2 3AT+5FOA.png|thumb|left|Selection strain without Zif268 with 3-AT and 5-FOA 8/12/11]] |
| - | [[File: 2011.08.12.Zif1 3AT+5FOA.png|thumb|none|Selection strain with Zif268 with 3-AT and 5-FOA 8/12/11]] | + | [[File: HARV2011.08.12.Zif1 3AT+5FOA.png|thumb|none|Selection strain with Zif268 with 3-AT and 5-FOA 8/12/11]]</div> |
| - | | + | <div id='816' style="display:none"> |
| - | | + | |
| | ==August 16== | | ==August 16== |
| | In prioritizing our work for the next two weeks, we decided to focus on three main experiments: | | In prioritizing our work for the next two weeks, we decided to focus on three main experiments: |
August 10
Team TolC
MAGE OZ052 and OZ123
- MAGE round 4a, time constants of 5.6s (OZ052) and 1.2s and 5.4s (OZ123). There appeared to be a spark for the OZ123 electroporation (Sarah)
Recombination of Zeocin into ECNR2
- Zeocin insert colonies appear to be growing extremely slowly, only 3 colonies after 14 hours, but may be due to rpoZ knockout
- Growing up culture in case we need to do lambda red of zeocin again
- PCR of the TolC, rpoZ and zeocin bands for colonies 1 and 2 off the zeocin plate.
- numbered, from 1-6, 1-3 is colony 1, 4-6 is colony 2. 1,4=TT 2,5=RR, 3,6=ZR
- PCR successful!! We have the real WN strain now. It should be ready for transformation tomorrow with the zif268 spec plasmid
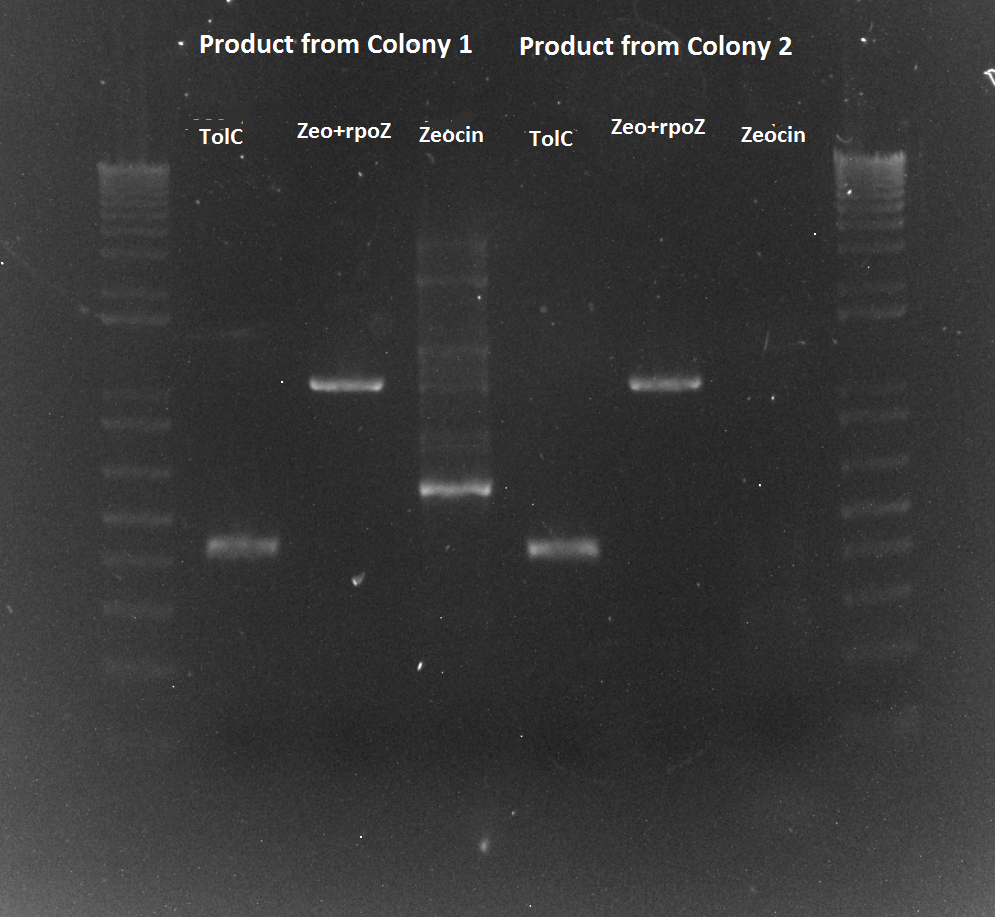
Zeocin/rpoZ and TolC PCR of lambda red colonies 8/10/11
Team Wolfe
Lambda Red RpoZ Knockout Plates
- Small colonies grew after about 14 hours on zeocin plates
Rpoz Knockout Check PCR
- Picked 24 colonies and used zeocin binding primers as a control and rpoZ binding primers to check for a knockout.
- 48 10µL reactions in total (24 colonies X 2 different primer sets mentioned above)
- Primers: rpoZ_R with Zeocin_R and rpoZ_R with rpoZ_F
- 64 degree annealing temperature
- 90 seconds elongation
Results
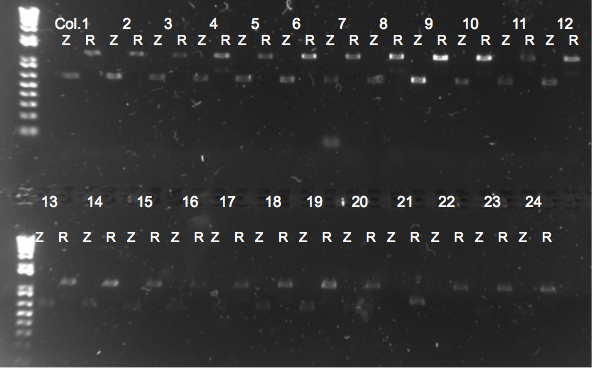
Zeocin/rpoZ PCR of lambda red colonies 8/10/11
- Reinoculated LB+zeo with culture 9 to use tomorrow for glycerol stocks and transformations
Digestion of pSB4K5 Plasmid
- Gel with yesterday's digestion product suggests that neither of the two restriction enzymes (Xba1 or Spe1) cut our plasmid. This was determined by comparing the band sizes to those obtained by midiporep of the pSB4K5 (whole) plasmid.
- Plan of action: We will use smaller reactions to increase the relative concentration of enzymes and we will use slightly more BSA buffer.
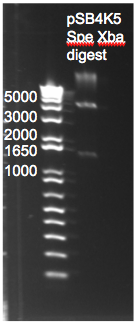
pSB4K5 digestion with Spe1 and Xba1
Today's Digestion of pSB4K5 Plasmid
- Xba1
- 16µL DNA (~1µg)
- 2µL buffer4
- .5µL BSA
- 1.5µL Xba1
- Spe1
- 16µL DNA (~1µg)
- 2µL buffer4
- .5µL BSA
- 1.5µL Spe1
- Xba1+Spe1
- 14.5µL DNA (~1µg)
- 2µL buffer4
- .5µL BSA
- 1.5µL Xba1
- 1.5µL Spe1
- Results: the size of the bands isn't what we expected (backbone should be ~3.4kb, insert ~1.2), but the comparison between the different digestions and the uncut plasmid does imply that the digestion we worked. We will go ahead and gel extract from the Spe+Xba digest and use both ligation and sequencing to be sure that this is indeed the plasmid we want.
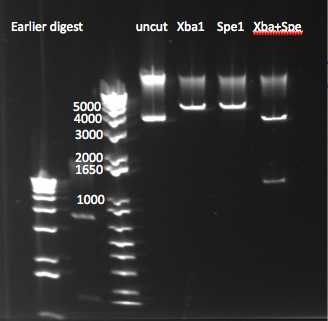
pSB4K5 digest with Spe1, Xba1, and both 8/10/11
Team ZF
Mini library ligation results
We checked our plates from our practice plate library assembly yesterday, and we got colonies!! Strangely, our 450 ul plate didn't have a huge number more colonies than the 50 ul plate. Dan suggested that this was because we plated all the cells in a 450 ul volume, which may have sloshed around and taken cells with it. Next time we should pellet the cells and resuspend in 100 ul and plate that. Based on our colony numbers with 50 ul, we should probably plate with 100 ul in the future.
Due to the proliferation of colonies, we picked 90 colonies with the assistance of Will. These 90 colonies should hopefully represent all 18 oligos in equal amounts. We did colony PCRs on all 90 with the junction PCR protocol, and sent them out for sequencing today. We saved these 90 colonies by adding LB/spec to each of the wells after suspending them in the 10 ul of water for the colony PCR. We will eventually harvest and gather all 18 assembled plasmids from this step.
Sequencing results for oligos 5 and 6
Sequencing indicated that most of our colonies worked, and that our F1s were inserted correctly! This is a huge step forward, and gives us good hopes that our 90 colonies also have their F1s inserted. Out of the 8 colonies each that we tested for oligo 5 and 6, only 5.8 (missing chunk on promoter), 5.3 (deletion on linker), and 6.8 (bad trace file) were bad. However, all the rest were good!
We also now have an idea of our combined ligation/transformation efficiency. NUMBERS HERE PLZ
Western blot to detect Zif268
Today we began a western blot in an attempt to detect Zif268 expression from IPTG induction. Due to the fact that the WB gels only have 10 lanes, we dropped the 2 uM IPTG sample for now, keeping the negative control (pze22g + gfp), 0 uM IPTG, 10 uM IPTG, and 100 uM IPTG.
Lysis buffer had the following composition:
- 150 mM NaCl
- 50 mM Tris-HCl
- 5 mM EDTA
- 1% Triton X-100
- 1x proteinase inhibitor
We used the following protocol for lysis:
- Spin down 3 ml of cells into a pellet, discarding the LB supernatant.
- Resuspend cells in 1.5 ml of ice cold lysis buffer, take 200 ul of this into a new tube and add 800 ul of cold lysis buffer.
- Sonicate at power level 2 in two sets of five bursts (each about one second long). Be sure to wait 10 seconds in between sets and keep tube on ice to prevent overheating.
- Spin down at maximum speed for 5 minutes at 4 degrees.
- Take supernatant into a new tube. This is the final protein lysis mixture.
Note! In the future, we should only spin down 200 ul of cells for lysis. According to Alain, most of the time they even just use 100 ul and directly boil it with SDS. May try this protocol tomorrow if current protocol doesn't work (the downside is that we have to start again from the culture stage and not the protein stage).
Prepping samples for the gel:
- For loading duplicates, load 9 ul of 5x sample buffer with 36 ul of protein for a total of 45 ul.
- Parafilm tubes to prevent popping, and boil for 5 minutes at 100 degrees.
- Load 10 ul of protein ladder into gel, and load 15 ul of sample into each well.
Note! For this particular gel, the wells go in this order from left to right: Marker, neg con, 0 uM, 10 uM, 100 uM, neg con, 0 uM, 10 uM, 100 uM, Marker.
Running samples on gel:
- Gel was run at 150 V until the marker band reached approximately 80% of the way to the bottom of the gel.
Transfer:
- Semi-dry transfer was accomplished by wetting blotting paper and nitrocellulose membrane in transfer buffer and making the sandwich on the transfer apparatus. From bottom to top (red to black electrode), the loading order was as follows:
- 2 sheets of blotting paper
- Nitrocellulose membrane
- Gel
- 2 sheets of blotting paper
- Transfer was run at 20 V for 1 hour.
Note! The side containing the protein is face up when the notch in the membrane is situated on the top right hand corner. However, due to troubles with the gel, the lanes are now in the opposite order of loading. So, with the protein right side up, the lanes go: marker, 100 uM, 10 uM, 0 uM, neg con, etc. Also, we forgot to roll bubbles out from the sandwich with the pipette fragment before transferring (my bad!) Hopefully this doesn't impact anything. One last note on the transfer: the current cap was 400 mA, and when we started the transfer, it initially began at 17 V and 400 mA. Over a few minutes, the voltage increased to 20 V and the current began to decrease, hitting 110 mA after a half hour of gradual decay, which is where we expected the current to be.
The transfer seemed to have worked, transferring the marker bands from the gel onto the membrane.
Blocking:
- Membrane was blocked at 4 degrees overnight.
Tomorrow we will proceed with primary and secondary antibody staining and band visualization.
Library ligation optimization
Dan suggests that we optimize the vector:insert ratio for our library assembly. We will try using the following insert ratios (assuming a vector unit of 1): 1, 2, 3, 4, 8, 10, 16. Unfortunately, today we did not have enough cut CB bottom plasmid to immediately begin the ligation and transformation, so we attempted to cut more CB bottom. After performing a cut with Bsa-HF using the previous protocol, we performed a PCR purification on the product, only to find that there were only 3.4 ng/ul of cut plasmid present. This is not enough to perform the ligation, and so we will have to try the CB bottom plasmid cut again tomorrow morning when the cultures have grown enough. We can then do a ligation and transformation tomorrow and count colonies on Friday.
As stated in the earlier section, we should probably plate 100 ul of transformed bacteria to standardize everything and to keep colony counting fairly manageable.
August 11th
Team Wolfe
Ligation of the HisUra Insert
- follow New England Biolabs protocol:
- 1.5µL of cut Spe ZFB-wp-hisura Xba insert (75ng,a 3-fold molar excess to backbone)
- 6µL cut pSB4K5 backbone (50ng)
- 1.5µL Water (bring volume up to 10µL)
- 1µL Quick T4 DNA Ligase
- 10µL 2X quick ligation buffer and mix
- spin briefly and incubate for 5 min at room temperature
- keep at -20˚C or on ice until transformation
- Transformation: ligation mixture cannot be directly electroporated, so we will start with top10 chem comp cells (see protocols page)
Transformation of pSB4K5 plasmid with His3Ura3 Insert into Chemically Competent Cells
- Transformed into delta HisB delta pyrF delta rpoZ Wolfe strain
- Cells plated in 1mL and 2mL quantities on Kan plates after 3 hours recovery
Transformation of Zif268 Plasmid
- Transformed into HisB delta pyrF delta rpoZ EcNR2 strain
- Cells were plated after 3.5 hours recovery on Spec plates at 10µL and 100µL
Transformation of the pSB4K5 Plasmid with His3Ura3 Insert into Electrocompetent Cells
- Transformed into delta HisB delta pyrF delta rpoZ Wolfe strain
- 3µL DNA from ligation reaction (unpurified)
Team ZF
Western Blot
PCR to get out GFP
Our new GFP forward primer arrived today, so we performed a PCR to get out the GFP region. Because the product will be used for gel extraction, we ran 4 reactions. We had an annealing temperature of ... and an extenstion time of 30 seconds.
Check oligo pool sequencing results
Unfortunately, most of the samples we sent in for sequencing had problems. For many, the primers did not anneal; for others there were poor quality reads. 13 samples in total had good quality reads. Among those we got a pretty good distribution among our 18 oligos. This is promising. We will perform a PCR on 2 random samples to ensure that the cross junction is present. If we get positive results, we will send in 24 more samples for sequencing.
Summary of sequencing results:
| Oligo
| Frequency
|
| 5 | 3
|
| 6 | 1
|
| 8 | 1
|
| 9 | 1
|
| 10 | 1
|
| 11 | 2
|
| 12 | 1
|
| 17 | 1
|
| 18 | 2
|
Finding the optimal backbone:insert for ligation
We wanted to find the optimal backbone to insert ratio for ligation. Previously we had successfully used 100 ng of backbone, and a ratio of 1 to 8. We decided to try the following ratios:
- 1:1, 1:2, 1:3, 1:4, 1:8, 1:10, 1:16
We ran out of cut CB Bottom backbone, so we needed to digest some more. We performed a miniprep on cultures that grew up overnight, using min-elute columns. Our final concentration of CB Bottom Plasmid was 50 ng/ul and 54 ng/ul.
We followed our usual protocol for digestion, and then performed a PCR purification, once again using a min-elute column to maximize our yield. However, the resulting product only had a concentration of ~5 ng/ul and a purity of ~1.6. We got similarly low yields yesterday, and are unsure where things are going wrong, since we have performed the same procedure in the past with success. Even if the plasmids weren't cutting, they should be present in the PCR purification product, so the low yields are puzzling. We decided to try a midi-prep tomorrow, so see if having more CB Bottom plasmids would help.
We grew up the following 100 mL cultures for midi-prep tomorrow: CB Bottom, Zif268.
Team TolC
Search for a better weak promoter
- We have searched for a better weak promoter than the one we have now and compared their sequences to the present weak promoter in the kan-ZFB-wp construct, sequences were not similar enough to change the weak promoter through MAGE
- To change the promoter would be a big process, so we are thinking of using a negative selection of TolC using colicin and then creating a higher TolC production standard to begin with which would allow the KS and KN growth curves separate more
Transformation of Zif268 Plasmid
- Performed transformation of the Zif268 plasmid into the good ECNR2+zeocin that definitely does have TolC
- Plated 1 mL of culture onto spec plate
Plater Reader
- Kept the IPTG concentration constant at 100 µM while changing the SDS concentration from 2% to 2.5%, 3%, 3.5%, and 4%
- Making two wells per different culture
- Cultures are changing by SDS and IPTG concentration, while also changing from KS, KN, WS, WN colonies
- Since colonies are not near mid-log we may have to fill plate reader tomorrow and run then as well
Gradient Plates
- Created 5 50 mL total gradient plates with 2x Kan in the wedge layer, 20ul of Bluejuice in each 25ml of LB+agar+Kan to mark it with a dye.
August 12th
Team Wolfe
Transformations
- EcNR2 strain with transformed zif268 plasmid grew on spec plates: After 15 hours, 1mL and 2mL plates have ~20 colonies each
- Wolfe strain with transformed ligated pSB4K5 plasmid had no growth on kan plate at 1mL and unusual looking growth on a tet plate at 1mL after 12 hours
- The pSB4K5 plasmid was successfully transformed into the chemically coMpetent cells - will midiprep these to obtain a clean sample of ligated pSB4K5 for another transformation in electrocompetent Wolfe strain cells.
PCR check for Zif268 plasmid insert
- Looks like the plasmid's presence is confirmed by PCR in 8 colonies
- Team ZF's "CJUN" protocol: pZE23G 2123R and pZE23G 3581F primers
- Note well 5 is fainter but is probably due to a loading error where a small volume of sample was left out.
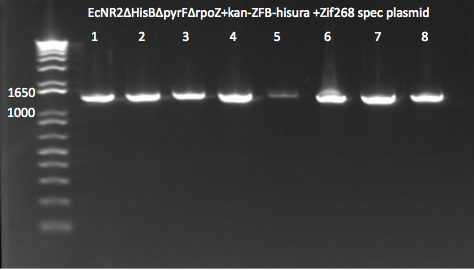
EcNR2 selection strain colonies transformed with zif268 8/12/11
Selection strain genotype confirmation
- We will do one last sequencing round of our EcNR2 selection strain to confirm that HisB and PyrF are indeed knocked out.
- Same parameters as previously described; His_R and PyrF_F used for sequencing
- PCR was successful, as shown by an E gel of several of the reactions.
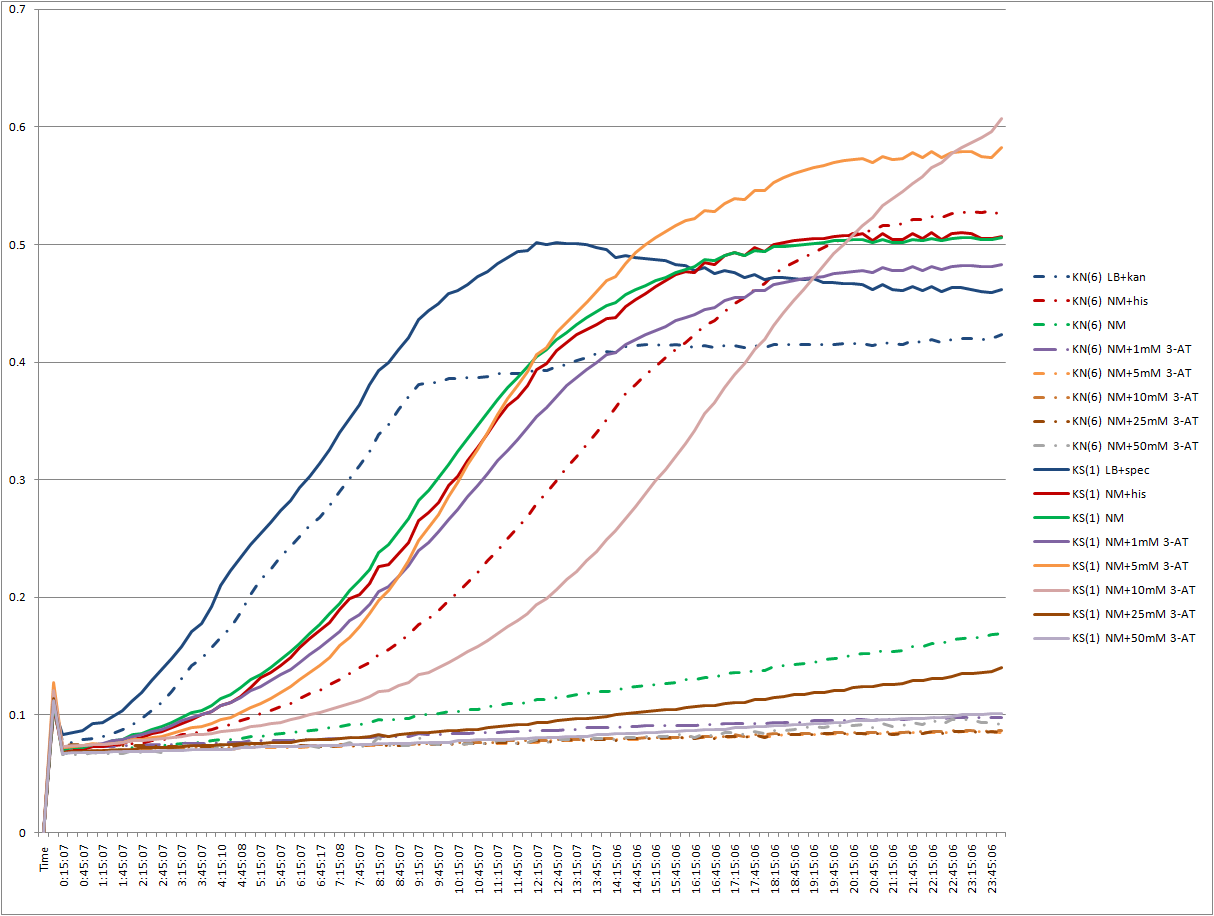
EcNR2 selection strain plate reader experiment
- We want to see how the EcNR2 selection strain works with and without plasmid, especially in the presence of various concentrations of 3-AT and 5-FOA.
- Wells 1, 14, 27, 40, 53: LB+kan/spec
- Wells 2, 15, 28, 41, 54: NM+his
- Wells 3, 16, 29, 42, 55: NM
- Wells 4, 17, 30, 43, 56: NM+1mM 3-AT
- Wells 5, 18, 31, 44, 57: NM+5mM 3-AT
- Wells 6, 19, 32, 45, 58: NM+10mM 3-AT
- Wells 7, 20, 33, 46, 59: NM+25mM 3-AT
- Wells 8, 21, 34, 47, 60: NM+50mM 3-AT
- Wells 9, 22, 35, 48, 61: NM+0.1mM 5-FOA
- Wells 10, 23, 36, 49, 62: NM+0.25mM 5-FOA
- Wells 11, 24, 37, 50, 63: NM+0.5mM 5-FOA
- Wells 12, 26, 38, 51, 64: NM+1mM 5-FOA
- Wells 13, 27, 39, 52, 65: NM+2.5mM 5-FOA
- Wells 1-13: empty
- Wells 14-26: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 (6)
- Wells 27-39: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 (9)
- Wells 40-52: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 +Zif268 spec (1)
- Wells 53-65: EcNR2∆HisB∆PyrF∆rpoZ+kan-ZFB-wp-his3-ura3 +Zif268 spec (2)
Team ZF
Midiprep of Zif268 and CB Bottom Plasmids
We performed a midi-prep on the Zif268 (whole) and CB Bottom plasmids. Zif268 was eluted in 100 ul of EB, while CB Bottom was eluted in 75 ul.
- Zif had a final concentration of 261.45 ng/ul and a 260/280 of 1.90.
- CB Bottom had a final concentration of 237.93 ng/ul and a 260/280 1.91.
Results of Cross Junction PCR of 2 random colonies from ligation
After running an e-gel we saw the expected 1.4 kb band. (The gel image is not available, because we were unable to get the software running at the time). We decided to proceed with sending 24 colonies out for sequencing.
Send out 24 colonies for sequencing
We performed a cross-junction PCR on the following colonies to send them out for sequencing:
- 1B, 1C, 1D, 1E, 1F, 1G, 1H, 2A, 3C, 3D, 3E, 3F, 3G, 3H, 4A, 4B, 10A, 10B, 10C, 10D, 10E, 10F, 10G, 10H
We picked cultures that did not have sequencing results from our previous attempt. A 1 ul of a 1:10 dilution of each culture was used as template in the PCR. We used the Junction Reverse primer.
Isothermal assembly and transformation of GFP reporter
We ran our PCR product from yesterday on a 1% agarose gel. We got an appropriate band at approximately 700 bp. (Unfortunately, we could not get the imaging software to work at the time, so no image is available)
We performed a gel extraction on our GFP PCR product, with the following yields:
- 114.90 ng/ul; 260/280 of 1.89
- 92.02 ng/ul; 260/280 of 1.94
We then performed an isothermal assembly, using the following recipe:
- 0.91 ul Zif 268 (backbone)
- 0.27 ul GFP
- 1.32 ul water
- 7.5 ul isothermal assembly mix
We transformed using ChemComp cells, plating 10 and 150 ul.
CB Bottom Digestion
We digested our CB Bottom plasmid, using our usual recipe. We ran the digestion for 2 hours at 37*, 20 minutes at 65*, and then let the reaction sit at 4* overnight.
August 16
In prioritizing our work for the next two weeks, we decided to focus on three main experiments:
- Preparing the oligo pool from the chip using Sri's protocol (using qPCR to amplify the pools equally)
- Continuing the experiment of picking out a positive hit amongst negative signal
- Biobricking our expression plasmids
Eventually, the first two experiments will come together in terms of actually using the chip and figuring out how to pull out and select the positive binders.
On average we expect about three students working at a time, so we'll have these three experiments running in parallel by each student. So this means our previous teams will have to be dissolved, also since TolC will no longer be used :'(.
Each experiment in more detail:
- We need to work out the protocol with Sri in order to prepare the oligo pools for ligation into our target plasmid backbones. This will involve running a qPCR to monitor the amount of amplification so that the oligos in each pool will be amplified in equal amounts (so that we get a representative pool of oligos that is not biased to one side or the other). We will need to do this with each subpool. Futhermore, we will have to confirm that the primers that Sri provided are indeed specific enough to pull out their target subpools only and not other subpools. This is important for the tweaking of our selection system, since we will need to know whether our selection system needs to be specific enough to pick out signal from 1:10,000 or 1:100,000.
- We will need to assemble a control practice library from the 18 practice plate oligos. We will basically formulate a negative control pool with equal amounts of each of the 18 oligos, and spike this pool with varying amounts of Zif268. We will then transform our selection strain with these pools with varying amounts of Zif268, and measure how specifically we can pick a winner out amongst non-winners (so instead of a bacteria dilution, it will be a plasmid dilution). We will use 8 different concentrations:
- Positive control (all Zif268)
- 1:10
- 1:100
- 1:1,000
- 1:10,000 (this is approximately the ratio we expect for a winner amongst a pool of 9,149 non-winners for the chip subpools)
- 1:100,000
- 1:1,000,000
- Negative control (only contains the 18 practice plate oligos)
- We will start figuring out the procedure for converting our expression plasmids into Biobricks for submission. This entails locating and eliminating all of the five specified restriction sites from our biobrick part (XbaI, SpeI, NotI, PstI, EcoRI), and assembling the part into a Biobrick submission plasmid (pSB1C3). This may be accomplished through the use of isothermal assembly to combine the Biobrick shipping plasmid backbone with our Biobrick part (the omega subunit + zinc finger array).
Sarah
TolC
TolC selection abandoned.
Wolfe
There appears to be low selection for the Wolfe strain as well. i.e. 1:10. One possibility is that the wrong strain was picked, the other. In the absence of the 96 well plate, we will use the culture tube, and possibly the 100 well, plate reader plate.
Team ZF
GFP reporter strain
We looked at the plate of GFP reporter that grew colonies and had the correct Shine-Dalgarno RBS with the correct spacer, putting it over the blue light of an e-gel box. We have glowing colonies! Only about seven of the ~250 colonies glowed, which may be due to an extra leaky promoter. We then picked a glowing colony and put it in the warm room to grow overnight in preparation for miniprep.
Control library assembly
We picked 24 colonies from our plate which was transformed with the library mix of the 18 practice plate oligos and grew them up in LB/spec for miniprep tomorrow.
 "
"