Team:Harvard/Template:NotebookData2
From 2011.igem.org
(Difference between revisions)
(→June 14 - Bioinformatics) |
(→June 14 - Wet Lab) |
||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<div id="614" style="display:none"> | <div id="614" style="display:none"> | ||
== June 14 == | == June 14 == | ||
| + | ===Wet Lab=== | ||
*Made four LB-based media solutions, and later created glycerol stocks from these and placed in -80⁰C freezer | *Made four LB-based media solutions, and later created glycerol stocks from these and placed in -80⁰C freezer | ||
**Selection strain (ΔHis3ΔPyrFΔrpoZ) in 3 mL LB and 3µL of 1000x Tet solution stock | **Selection strain (ΔHis3ΔPyrFΔrpoZ) in 3 mL LB and 3µL of 1000x Tet solution stock | ||
| Line 54: | Line 55: | ||
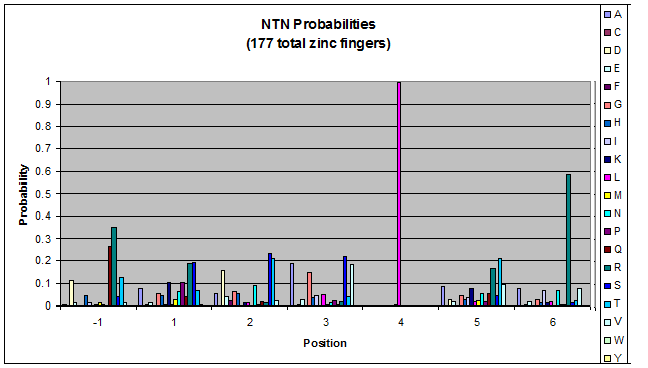
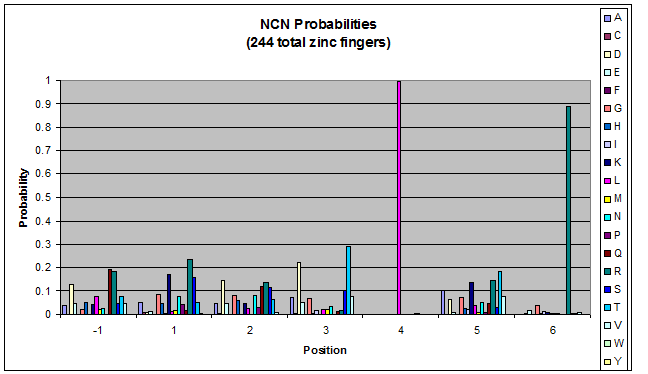
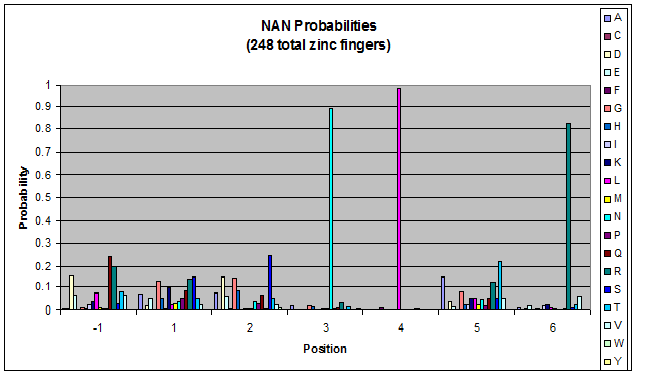
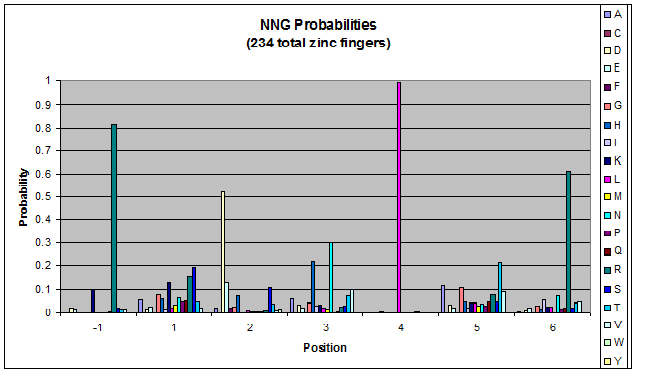
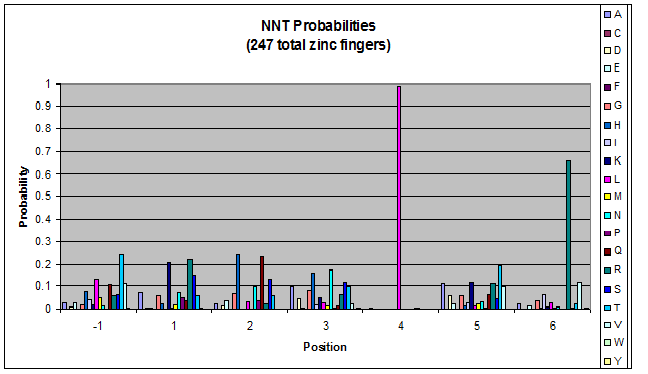
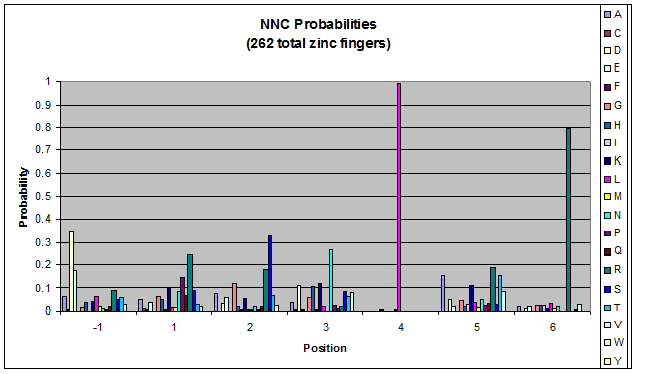
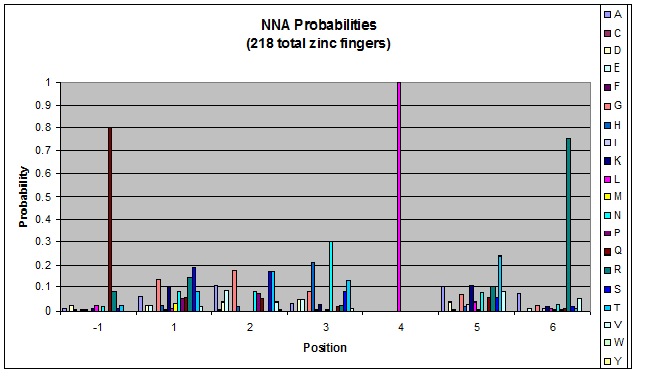
We finished writing the generate function, and now have a working sequence generator. We also began more in-depth research into the 2011 work by Persikov which deals with how the zinc finger binds to DNA. He predicts several relations which we should be able to test. | We finished writing the generate function, and now have a working sequence generator. We also began more in-depth research into the 2011 work by Persikov which deals with how the zinc finger binds to DNA. He predicts several relations which we should be able to test. | ||
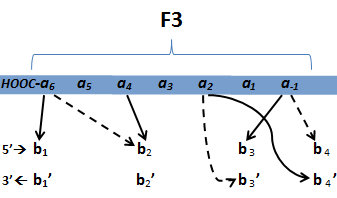
| - | [[File:HARVInteraction Map.png|frame|right|Proposed novel interactions between helical zinc finger residues and base pairs of the target DNA sequence (based on Persikov 2011 | + | [[File:HARVInteraction Map.png|frame|right|Proposed novel interactions between helical zinc finger residues and base pairs of the target DNA sequence (based on [http://www.ncbi.nlm.nih.gov/pubmed/21572177 Persikov 2011])]] |
Persikov sent us his SVM code (used to calculate the probability of a sequence binding to given DNA), so we also worked on adapting this to use when narrowing our sequences to those most likely to work. | Persikov sent us his SVM code (used to calculate the probability of a sequence binding to given DNA), so we also worked on adapting this to use when narrowing our sequences to those most likely to work. | ||
| Line 62: | Line 63: | ||
**Persikov uses the information between these four amino acid-base interactions in his SVM, to determine whether a finger would be a good binder to a particular DNA sequence. | **Persikov uses the information between these four amino acid-base interactions in his SVM, to determine whether a finger would be a good binder to a particular DNA sequence. | ||
*In order to use his program, we need to convert the helices that are created by the generator into a format the SVM accepts. | *In order to use his program, we need to convert the helices that are created by the generator into a format the SVM accepts. | ||
| - | **The SVM only considers the four canonical interactions. It assigns a numerical value to each possible amino acid-base combination. The program that converts our data into a format the SVM accepts creates a string with these numerical values based on Persikov's key. (See [ | + | **The SVM only considers the four canonical interactions. It assigns a numerical value to each possible amino acid-base combination. The program that converts our data into a format the SVM accepts creates a string with these numerical values based on Persikov's key. (See [https://2011.igem.org/Team:Harvard/Brainstorming#Persikov_2009_SVM this page] for more details on this program.) |
| Line 68: | Line 69: | ||
<div id="615" style="display:none"> | <div id="615" style="display:none"> | ||
| - | ==June 15th== | + | ==June 15th - Wet Lab== |
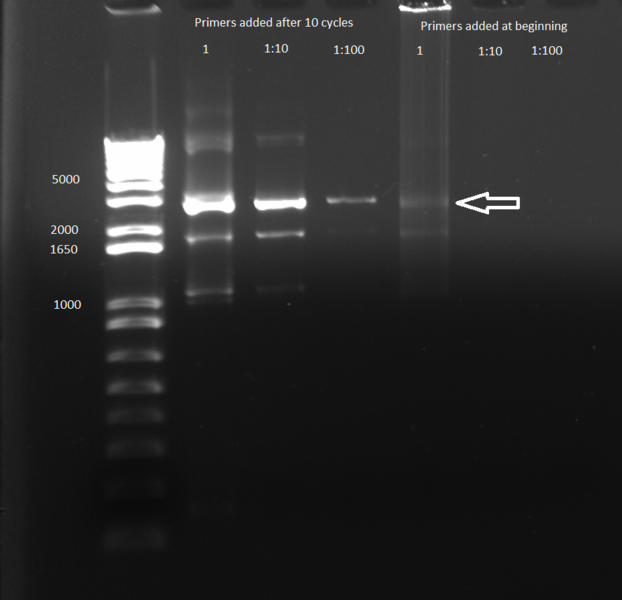
'''Gradient PCR:''' 5µL of each PCR product were run on a 1% gel. No bands appeared: the PCR appears to not have worked. | '''Gradient PCR:''' 5µL of each PCR product were run on a 1% gel. No bands appeared: the PCR appears to not have worked. | ||
| Line 147: | Line 148: | ||
*Justin continued work on a sequence-finding program, the most up to date version can be found in the Dropbox under code/zfsitefinder.html. | *Justin continued work on a sequence-finding program, the most up to date version can be found in the Dropbox under code/zfsitefinder.html. | ||
| - | *Justin and Will found 10 candidate sequences across 4 diseases that hopefully should encompass a good amount of diversity in terms of expanding the ZF library. These sequences can be found in the table below, with more details [ | + | *Justin and Will found 10 candidate sequences across 4 diseases that hopefully should encompass a good amount of diversity in terms of expanding the ZF library. These sequences can be found in the table below, with more details [https://2011.igem.org/File:HARVZF_Binding_Sequence_Candidates.xls here]. |
{| class="wikitable" cellpadding="5" | {| class="wikitable" cellpadding="5" | ||
| Line 171: | Line 172: | ||
|} | |} | ||
| - | *We collected 15 alternative zinc finger backbones (different from zif268 backbone) and their corresponding base sequences. Many of these were from Persikov 2011 and all binding sequences were confirmed on the | + | *We collected 15 alternative zinc finger backbones (different from zif268 backbone) and their corresponding base sequences. Many of these were from Persikov 2011 and all binding sequences were confirmed on the [http://www.pdb.org/pdb/home/home.do Protein Data Bank website]. The zinc finger PDB ID's and related links are: |
| Line 241: | Line 242: | ||
|}</div> | |}</div> | ||
<div id="616" style="display:none"> | <div id="616" style="display:none"> | ||
| - | |||
| - | * | + | ==June 16th - Wet Lab== |
| + | |||
| + | *There was totally a crazy bee hive outside today!! | ||
'''Glycerol Stock pKD42''' | '''Glycerol Stock pKD42''' | ||
| Line 387: | Line 389: | ||
***We can now create lists of the zinc fingers that bind to any triplet, and create interaction matrices and frequency tables for any triplet input.</div> | ***We can now create lists of the zinc fingers that bind to any triplet, and create interaction matrices and frequency tables for any triplet input.</div> | ||
<div id="617" style="display:none"> | <div id="617" style="display:none"> | ||
| - | ==June 17th== | + | ==June 17th - Wet Lab== |
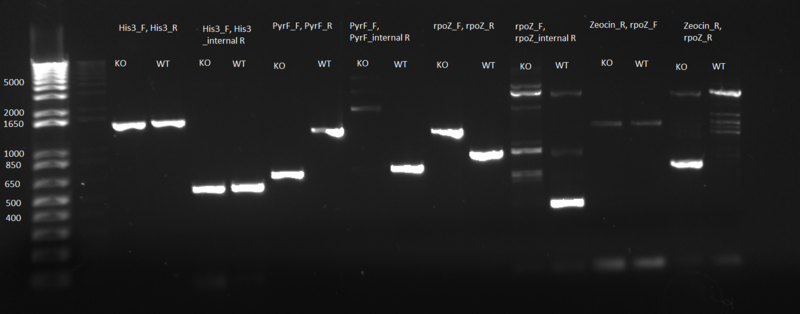
'''Update on selection strain knockout status:''' We are trying to reach Addgene to check how His3 was knocked out---instead of deleting the gene, they may have simply introduced an early stop codon. If that's the case, our gel would have the correct bands because the primers we designed can only show whether a deletion or insertion was in that locus. | '''Update on selection strain knockout status:''' We are trying to reach Addgene to check how His3 was knocked out---instead of deleting the gene, they may have simply introduced an early stop codon. If that's the case, our gel would have the correct bands because the primers we designed can only show whether a deletion or insertion was in that locus. | ||
| Line 417: | Line 419: | ||
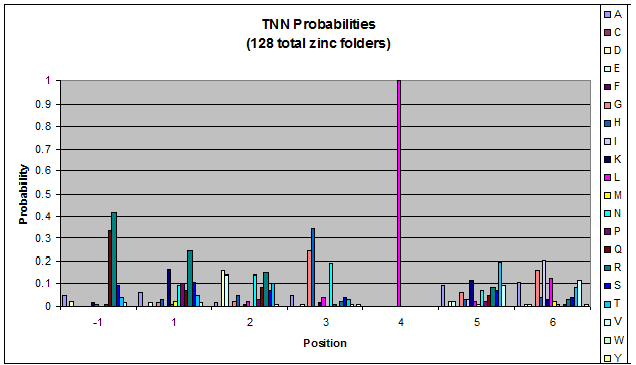
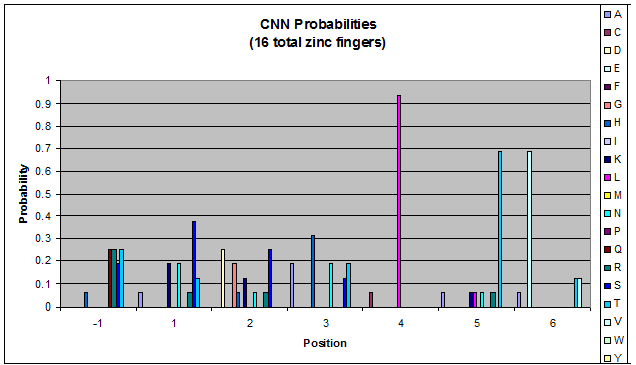
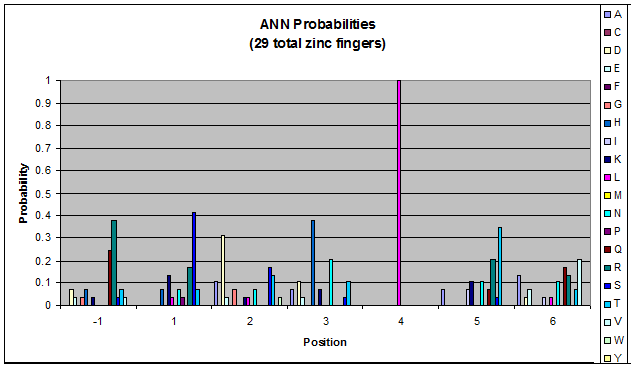
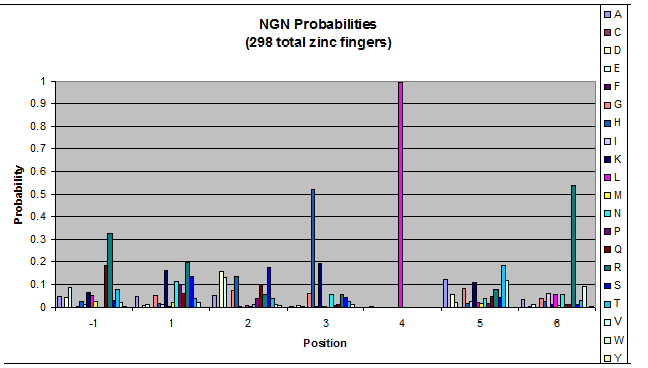
***Graph: # of var (# of tries by the computer) vs. % space covered | ***Graph: # of var (# of tries by the computer) vs. % space covered | ||
| - | [[File:HARVInteraction Map.png|frame|right|Proposed interactions between helical zinc finger residues and base pairs of the target DNA sequence (based on Persikov 2011 | + | [[File:HARVInteraction Map.png|frame|right|Proposed interactions between helical zinc finger residues and base pairs of the target DNA sequence (based on [http://www.ncbi.nlm.nih.gov/pubmed/21572177 Persikov 2011])]] |
====Options for Target DNA Sequences / ZF Helices==== | ====Options for Target DNA Sequences / ZF Helices==== | ||
| Line 439: | Line 441: | ||
---- | ---- | ||
====References==== | ====References==== | ||
| - | |||
#Persikov2011 pmid=21572177 | #Persikov2011 pmid=21572177 | ||
#CodonUsage http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/in-vitro-genetics/codon-usage.html | #CodonUsage http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/topics/in-vitro-genetics/codon-usage.html | ||
#OpenWetWareCodonUsage http://openwetware.org/wiki/Escherichia_coli/Codon_usage | #OpenWetWareCodonUsage http://openwetware.org/wiki/Escherichia_coli/Codon_usage | ||
#NIHRareCodonCalculator http://nihserver.mbi.ucla.edu/RACC/ | #NIHRareCodonCalculator http://nihserver.mbi.ucla.edu/RACC/ | ||
| - | + | </div> | |
<div id="620" style="display:none"> | <div id="620" style="display:none"> | ||
| - | ==June 20th== | + | ==June 20th - Wet Lab== |
*Grew up colony of the selection strain with pKD46 in an attempt to reach mid-log and create glycerol stock | *Grew up colony of the selection strain with pKD46 in an attempt to reach mid-log and create glycerol stock | ||
 "
"