Team:Glasgow/Modeling
From 2011.igem.org
HannahRalph (Talk | contribs) |
|||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | { | + | {{Team:Glasgow/Header}} |
| - | + | <html> | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <center><h1><b>Mathematical model guide engineering of biofilms</b></h1></center> |
| - | <a | + | <center><img src="https://static.igem.org/mediawiki/2011/3/35/M-latherin2.gif"></center> |
| + | |||
| + | <h4>Modelling aims</h4><p> | ||
| + | To guide the engineering of our biofilm system, we had to apply a mathematical modelling strategy. For this, quantitative biophysical models of our system were created. The main aims of these models were to predict the resolution and to estimate the rate of biofilm dispersal. This determines the spatial resolution of our light-controlled biofilms and is central for engineering their functionality. This was done by predicting the time required for creation of a specific protein, the rate at which it diffuses through biofilm and how long it takes to generate an effect by attaining a critical concentration threshold. | ||
| + | <p> | ||
| + | Three separate methods were devised for the purpose of biofilm dispersal: expression of a surfactant, reduction in levels of cyclic diguanylate monophosphate (c-di-GMP) and selective cell lysis. Surfactants should aid dispersal of a biofilm by reducing friction in its structure. This prediction is based on biofilm dispersal previously measured through expression of surfactin. [Mireles et al., 2001] | ||
| + | <p> | ||
| + | C-di-GMP is essential for biofilm formation. Research suggests [Kaplan, J., 2010] that decreasing the intracellular concentration of this molecule will result in increased motility and decreased extracellular matrix formation – thus triggering biofilm dispersal. | ||
| + | <p> | ||
| + | The aim of the iGEM team is to engineer a system where each dispersal mechanism triggered by a light sensitive promoter responsive to a specific wavelength of light. The models are designed to predict the resolution of this selective dispersal mechanism over a period of time following initial activation. | ||
| + | <p> | ||
| + | Cell death is expected to trigger biofilm dispersal because its structure is weakened by the absence of bacteria and also exopolysaccharide production is obstructed. Moreover, cell lysis in the biofilm causes adjacent cells to increase motility, thus promotes biofilm dispersal [Kaplan, J., 2010; Webb et al., 2003]. | ||
| + | <p> | ||
| + | Models were generated including quantitative descriptions of the behaviour of the following proteins: (* indicates novel biobrick)<br> | ||
| + | <b>*Latherin:</b> a surfactant protein component of horse sweat. | ||
| + | <br><b>*Ranaspumin-2:</b> a surfactant protein used in the formation of foam nests built by the Tangara frog. | ||
| + | <br><b>*Phosphodiesterase with VieA domain (PDE):</b> a protein that regulates c-di-GMP levels through cleavage of ester bonds. The VieA domain is specific to c-di-GMP. | ||
| + | <br><b>Colicin E2 (BBa_K131000):</b> a DNase which triggers cell lysis. | ||
| + | <br><b>T4 endolysin (BBa_K112806):</b> a lysozyme-like protein produced by T4 bacteriophage in order to lyse the cell and release virions. | ||
| + | |||
| + | <h4>Translation</h4> | ||
| + | Translation rate calculations were based on average translation rates of E. coli, the following assumptions were made: | ||
| + | <br> - RNA transcripts of the protein, once triggered, were generated at an average rate of 40 aa/sec | ||
| + | <br> - each cell contained an average number of ribosomes – 18000 ribosomes/cell | ||
| + | <p> | ||
| + | Using these assumptions, the number of ribosomes working on a single RNA transcript was estimated. The rate of protein translation was predicted based on the length of the protein being produced. Also, expression of those molecules is driven by the same ribosome binding site (RBS). | ||
| + | <p> | ||
| + | <table border><tr><b>Table 1. Details on molecules used in diffusion models</b></tr> | ||
| + | <tr><td>Name of molecule</td> | ||
| + | <td>Length (a.a.)</td> | ||
| + | <td>Molecular weight (g/mol)</td> | ||
| + | <td>Diffusion rate (mm<sup>2</sup>/min)</td> | ||
| + | <td>Critical concentration (molecules)</td></tr> | ||
| + | <tr><td>Ranaspumin-2</td> | ||
| + | <td>98</td> | ||
| + | <td>11287</td> | ||
| + | <td>1.68*10<sup>-4</sup></td> | ||
| + | <td>374</td></tr> | ||
| + | <tr><td>T4 endolysin</td> | ||
| + | <td>164</td> | ||
| + | <td>18500</td> | ||
| + | <td>1.52*10<sup>-4</sup></td> | ||
| + | <td>3000</td></tr> | ||
| + | <tr><td>Latherin</td> | ||
| + | <td>220</td> | ||
| + | <td>24400</td> | ||
| + | <td>1.38*10<sup>-4</sup></td> | ||
| + | <td>173</td></tr> | ||
| + | <tr><td>Colicin E2</td> | ||
| + | <td>563</td> | ||
| + | <td>60000</td> | ||
| + | <td>0.72*10<sup>-4</sup></td> | ||
| + | <td>1629</td></tr></table> | ||
| + | <p> | ||
| + | <h4>Diffusion</h4> | ||
| + | Specific parameters for all of the molecules were initially searched for in the literature (possible diffusion coefficients, size, translation rates and critical concentrations). Due to the lack of specific information and experimentation done on certain molecules, in most cases it was necessary to make additional assumptions and/or use average values to make reasonable estimates of the parameter values operating our system. | ||
| + | <p> | ||
| + | The diffusion rate depends on the size of the molecule and the viscosity of the environment. Biofilm is usually very dense and viscous, thus diffusion through it is impaired and slow. We used a one-dimensional diffusion equation to calculate the rate at which modelled molecules should move through the biofilm:<br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2011/4/41/Equation1.jpg"></center> | ||
| + | <p align="right">Equation 1</p> | ||
| + | <p> | ||
| + | where:<br> P (r,t) – number of molecules at the certain distance after a certain time | ||
| + | <br>A – total number of molecules in the system | ||
| + | <br>r – distance from point of origin (mm) | ||
| + | <br>D – diffusion coefficient | ||
| + | <br>t – time (min) | ||
| + | <p> | ||
| + | Based on the research of Lawrence et al. (1994) and assuming that the diffusion coefficient depends on the molecular weight of the molecule in a linear fashion, we estimated possible biofilm diffusion rates for Latherin, Ranaspumin-2, T4 endolysin and Colicin E2 (Table 1). | ||
| + | <p> | ||
| + | For PDE the purpose of the model was to estimate how long it would take to deplete the cell completely of c-di-GMP after expression. It was assumed that each cell contained an average concentration of c-di-GMP and the enzyme was working at its full capacity. | ||
| + | <p> | ||
| + | Required calculations and graph plotting were performed with OpenOffice Spreadsheet and GnuPlot software. | ||
| + | <p> | ||
| + | The results from models are very informative for guiding our biological engineering of functionalised biofilms. In the case of Colicin E2, research shows [Pugsley, A., 1983] that the protein needs to be expressed for about 90 minutes to cause the cells to lyse. However, molecules like latherin and ranaspumin-2, because of their mode of action, are expected to require at least an hour to cause a desirable effect. For that reason the initial concentrations need to exceed the critical concentrations by a factor 8, so that it will not be diluted by diffusion before being able to affect the area we want to sculpt through the dispersion of biofilm. Since the time of expression of the relevant molecules is controlled by light inducible promoters in our engineered biofilms, concentrations will depend on the timing of the respective light exposure. Estimated times and concentrations are presented in Table 2. | ||
| + | <p> | ||
| + | <table border><tr><b>Table 2. Calculated times of light exposure required to achieve necessary concentrations of the molecules</b></tr> | ||
| + | <tr><td>Name of molecule</td> | ||
| + | <td>Translation rate (molecule/min)</td> | ||
| + | <td>Achieving Critical Concentration (min)</td> | ||
| + | <td>Achieving 8xCritical concentration (min)</td></tr> | ||
| + | <tr><td>Colicin E2</td> | ||
| + | <td>18.3</td> | ||
| + | <td>90</td> | ||
| + | <td>N/A</td></tr> | ||
| + | <tr><td>T4 endolysin</td> | ||
| + | <td>57.2</td> | ||
| + | <td>52</td> | ||
| + | <td>N/A</td></tr> | ||
| + | <tr><td>Latherin</td> | ||
| + | <td>46.9</td> | ||
| + | <td>3</td> | ||
| + | <td>29</td></tr> | ||
| + | <tr><td>Ranaspumin-2</td> | ||
| + | <td>105.3</td> | ||
| + | <td>3</td> | ||
| + | <td>28</td></tr></table> | ||
| + | <p> | ||
| + | The attainable spatial resolution was calculated based on the diffusion rates of the molecules, i.e. what will be the size of the affected area compared with the area being illuminated. The values of achievable resolution was acquired by calculating at what distance from the initial point we can still observe a concentration, achieved through the diffusion, exceeding the critical concentration for the respective molecule. The initial calculations show that we should be able to achieve an average resolutions as small as ~0.1 mm. The values are presented in Table 3. | ||
| + | |||
| + | <table border><tr><b>Table 3. The attainable spatial resolution calculated with the use of the single-cell models for Latherin, Ranaspumin-2, Colicin E2 and T4 endolysin.</b></tr> | ||
| + | <tr><td>Name of the molecule</td> | ||
| + | <td>The attainable spatial resolution (mm)</td> | ||
| + | <td>The spatial resolution in terms of No. cells</td></tr> | ||
| + | <tr><td>Latherin</td> | ||
| + | <td>0.08</td><td>40</td></tr> | ||
| + | <tr><td>Ranaspumin-2</td> | ||
| + | <td>0.09</td><td>45</td></tr> | ||
| + | <tr><td>Colicin E2</td> | ||
| + | <td>N/A</td><td>1</td></tr> | ||
| + | <tr><td>T4 Endolysin</td> | ||
| + | <td>N/A</td><td>1</td></tr></table> | ||
| + | |||
| + | <p> | ||
| + | However, while estimating resolution the protein degradation rate should also be taken into consideration. Where we do this, the number of molecules has to be modelled as being time-dependent according to the following equation:<br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2011/3/38/Equation2.jpg"></center> | ||
| + | |||
| + | <p align="right">Equation 2</p> | ||
| + | <p> | ||
| + | where:<br> P<sub>f</sub> (r,t) – Final number of molecules at given distance at given time | ||
| + | <br>P (r,t) – initial number of molecules according to Equation 1 | ||
| + | <br>λ – rate of degradation (molecule/min) | ||
| + | <br>t – time passed (min) | ||
| + | <p> | ||
| + | At the moment specific rates of degradation of the molecules in our system are not known, so simulations had to be done for estimated parameters based on general protein degradation rates. In general, our models predict that resolutions will be even better than 0.1 mm, when diffusing molecules are also degraded. Graphical representations of models of diffusion through biofilm for relevant molecules are shown in Figures 1 and 2. All three molecules are shown on a single Figure 3 to allow comparison of their relative diffusion rate through a biofilm. | ||
| + | <p> | ||
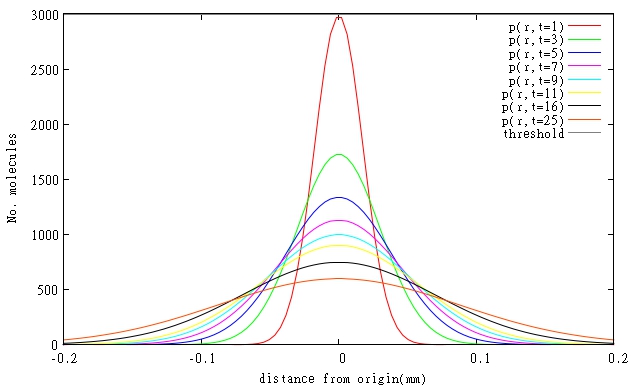
| + | <table><tr><b>Figure.1. Relative One-dimensional Diffusion of (a)Latherin, (b)Ranaspumin-2, (c)Colicin E2 and (d)T4 Endolysin at certain points in time, when initial concentrations of Latherin and Ranaspumin-2 are 8x critical concentration. Threshold indicates critical concentration required for the molecule to work.</b></tr> | ||
| + | |||
| + | <tr><td>(a) Latherin</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/e/ef/Lather2d.jpg"></td></tr> | ||
| + | <tr><td>(b) Ranaspumin-2</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/4/46/Ranaspumin2d.jpg"></td></tr> | ||
| + | <tr><td>(c) Colicin E2</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/6/62/Colicin2d.jpg"></td></tr> | ||
| + | <tr><td>(d) T4 Endolysin</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/1/1c/T42d.jpg"></td></tr></table> | ||
| + | |||
| + | <p> | ||
| + | <table><tr><b>Figure.2. Relative One-dimensional diffusion of (a)Latherin, (b)Ranaspumin-2, (c)Colicin E2 and (d)T4 Endolysin within first 60 min, when initial concentrations of Latherin and Ranaspumin-2 are 8x critical concentration.</b></tr> | ||
| + | <tr><td>(a) Latherin</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/f/f7/Latherin3d.jpg"></td></tr> | ||
| + | <tr><td>(b) Ranaspumin-2</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/7/79/Ranaspumin3d.jpg"></td></tr> | ||
| + | <tr><td>(c) Colicin E2</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/7/7d/Colicine23d.jpg"></td></tr> | ||
| + | <tr><td>(d) T4 Endolysin</td></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/3/3c/T43d.jpg"></td></tr></table> | ||
| + | |||
| + | <p> | ||
| + | <table><tr><b>Figure.3. Comparison of rate of diffusion through a biofilm of Colicin E2, Ranaspumin-2 and Latherin after 1 minute from start of the diffusion, when initial concentration of Ranaspumin-2 and Latherin is 4xCritical concentrations.</b></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/7/77/Col-rana-lath-2d.jpg"></td></tr></table> | ||
| + | |||
| + | <p> | ||
| + | For the model of PDE activity we made several simple calculations and assumptions. The concentration of c-di-GMP in an average bacterial cell is low, 1.25x10<sup>-17</sup> [μmol/cell], the Vmax of PDE is 1.5457x10<sup>-15</sup> [μmol min<sup>-1</sup> molecule<sup>-1</sup>]. This means an average cell can produce around 99.6 molecules of PDE per minute. As a result, fully active PDE should be able to deplete a cell of c-di-GMP in ~0.01 minutes (0.6 secs). Therefore, to compensate for any inaccuracies, allowing production of PDE for a full minute should deprive the cell completely of c-di-GMP. Lack of c-di-GMP would increase motility of bacteria and cause biofilm dispersal. Using this in concordance with surfactant protein expression should generate a cumulative effect on dispersal. | ||
| + | <p> | ||
| + | All above models were calculated for a single cell. Because of that and also the fact that for building the models many assumptions and average values were used, the properties of our real engineered system will be slightly different from the predictions shown above. In order to improve the accuracy of our predictions, additional quantitative experiments have to be performed to obtain more precise and specific parameter values. These could then be used to improve the model accuracy for future predictions. | ||
| + | <h4>Multi-cell models</h4> | ||
| + | <p> | ||
| + | Due to the fact that an average biofilm comprises of 3x10<sup>7</sup> CFU/cm<sup>2</sup> and an average E. coli is 2 μm long, the density of cell in any biofilm will be relatively high. Therefore, in order to predict the behaviour of our system when it involves more than one cell, the following models for Latherin and Colicin E2 were created. Both of those models consider a system where there are 21 cells in a row the centres of which are spaced at a distance of 2 μm between each other. Because they all produce the same protein, the overall concentration in the environment is higher than in the single cell situation. That causes the spatial resolution to be poorer, since the concentration exceeds the critical concentration farther from the point of origin than in the single cell models. The difference between the single-cell and the multi-cell models varies, depending on the rate of diffusion and the critical concentrations, but in all cases it is affected. However, a better resolution can be easily acquired by reducing the time the cells are exposed to the specific light wavelength, hence the production of the relevant proteins will be smaller. Therefore, the concentration will not exceed the critical concentration in areas far away from the illuminated parts. That means that the spatial resolution can be precisely controlled by time of light exposure. To establish the optimal time of illumination, a series of experiments need to be conducted. Figures 4 and 5 show the graphical representations of those models. | ||
| + | <p> | ||
| + | <table><tr><b>Figure 4. Model of Latherin where 21 cells are next to each other in a row. Two plots are shown: the first shows cells illuminated for 29 min and the second shows cells illuminated for 15 min. Plots show concentration of the protein after 360 min of diffusion. Threshold shows the critical concentration needed for Latherin to work.</b></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/2/2a/Latherin21c-comparison.jpg"></td></tr></table> | ||
| + | |||
| + | <p> | ||
| + | <table><tr><b>Figure 5. Model of Colicin E2 where 21 cells are next to each other in a row. Two plots are shown: the first shows cells illuminated for 90 min, the second shows cells illuminated for 45 min. Plots show concentration of the protein after 70 min of diffusion. Threshold indicates the critical concentration needed by Colicin E2 to lyse a cell.</b></tr> | ||
| + | <tr><td><img src="https://static.igem.org/mediawiki/2011/7/71/Colicin21c-comp.jpg"></td></tr></table> | ||
| + | |||
| + | <h4>References:</h4> | ||
| + | 1. Beeley et al., 1986. Isolation and characterization of latherin, a surface-active protein from horse sweat. Biochemical Journal, 235, pp. 645-650.<br> | ||
| + | 2. Buchler et al., 2005. Nonlinear protein degradation and the function of genetic circuits. PNAS, 102(27), pp. 9559-9564.<br> | ||
| + | 3. <i>Escherichia coli</i> statistics. Institute for Biomolecular Design. Source: internet | ||
| + | [http://www.ccdb.ualberta.ca/CCDB/cgi-bin/STAT_NEW.cgi].<br> | ||
| + | 4. Goldberg, A. & Dice, J., 1974. Intracellular protein degradation in mammalian and bacterial cells. Annual Review of Biochemistry, 43, 835-869.<br> | ||
| + | 5. Kennedy, M., 2011. Latherin and other biocompatible surfactant proteins. Biochemical Society Transactions, 39, pp. 1017-1022.<br> | ||
| + | 6. Kaplan, J., 2010. Biofilm dispersal: Mechanisms, Clinical Implications, and Potential Therapeutic Uses. Journal of Dental Research, 89(3), pp.205-218.<br> | ||
| + | 7. Konisky, J., 1982. Colicins and other bacteriocins with established modes of action. Annual Review of Microbiology, 36, pp. 125-144.<br> | ||
| + | 8. Lawrence et al., 1994. Determination of diffusion coefficients in biofilms by confocal laser microscopy. Applied and Environmental Microbiology, 60(4), pp. 1166-1173.<br> | ||
| + | 9. Levi et al., 2011. Cyclic Diguanylate Signalling Proteins Control Intracellular Growth of Legionella pneumophila. mBio, 2(1), e00316-10.<br> | ||
| + | 10. Jackson et al., 2001. Growing reproducible biofilms with respect to structure and viable cell counts. Journal of Microbiological Methods, 47, pp. 1-10.<br> | ||
| + | 11. Mackenzie et al., 2009. Ranaspumin-2: structure and function of a surfactant protein from the foam nests of a tropical frog. Biophysical Journal, 96, pp. 4984-4992.<br> | ||
| + | 12. Mireles et al., 2001. Salmonella enterica Serovar Typhimurium Swarming mutants with altered biofilm-forming abilities: Surfactin inhibits biofilm formation. Journal of Bacteriology, 183(20), pp. 5848-5854.<br> | ||
| + | 13. Molecular structures. RCSB Protein Data Bank. Source: internet | ||
| + | [http://www.pdb.org/pdb/home/home.do]<br> | ||
| + | 14. Pugsley, A., 1983. Obligatory coupling of Colicin release and lysis in Mitomycin-treated Col+ Escherichia coli. Journal of General Microbiology, 129, pp. 1921-1928.<br> | ||
| + | 15. Tamayo et al., 2005. The EAL domain Protein VieA is a cyclic diguanylate phosphodiesterase. The Journal of Biological Chemistry, 280(39), pp. 33324-33330.<br> | ||
| + | 16. Ursell, T., 2005. Diffusion of solid particles confined in viscous fluid. Source: internet | ||
| + | [http://www.rpgroup.caltech.edu/courses/aph162/2006/Protocols/diffusion.pdf].<br> | ||
| + | 17. Ursell, T., 2007. The diffusion equation: a multi-dimensional tutorial. Source: internet | ||
| + | [http://www.rpgroup.caltech.edu/~natsirt/aph162/diffusion.pdf].<br> | ||
| + | 18. Wang et al., 2000. Holins: the protein clocks of bacteriophage infections. Annual Review of Microbiology, 54, pp. 799-825.<br> | ||
| + | 19. Webb et al., 2003. Cell death in Pseudomonas aeruginosa Biofilm development. Journal of Bacteriology, 185(15), pp. 4585-4592. | ||
| + | <br></html> | ||
Latest revision as of 15:18, 20 September 2011

Mathematical model guide engineering of biofilms

Modelling aims
To guide the engineering of our biofilm system, we had to apply a mathematical modelling strategy. For this, quantitative biophysical models of our system were created. The main aims of these models were to predict the resolution and to estimate the rate of biofilm dispersal. This determines the spatial resolution of our light-controlled biofilms and is central for engineering their functionality. This was done by predicting the time required for creation of a specific protein, the rate at which it diffuses through biofilm and how long it takes to generate an effect by attaining a critical concentration threshold.
Three separate methods were devised for the purpose of biofilm dispersal: expression of a surfactant, reduction in levels of cyclic diguanylate monophosphate (c-di-GMP) and selective cell lysis. Surfactants should aid dispersal of a biofilm by reducing friction in its structure. This prediction is based on biofilm dispersal previously measured through expression of surfactin. [Mireles et al., 2001]
C-di-GMP is essential for biofilm formation. Research suggests [Kaplan, J., 2010] that decreasing the intracellular concentration of this molecule will result in increased motility and decreased extracellular matrix formation – thus triggering biofilm dispersal.
The aim of the iGEM team is to engineer a system where each dispersal mechanism triggered by a light sensitive promoter responsive to a specific wavelength of light. The models are designed to predict the resolution of this selective dispersal mechanism over a period of time following initial activation.
Cell death is expected to trigger biofilm dispersal because its structure is weakened by the absence of bacteria and also exopolysaccharide production is obstructed. Moreover, cell lysis in the biofilm causes adjacent cells to increase motility, thus promotes biofilm dispersal [Kaplan, J., 2010; Webb et al., 2003].
Models were generated including quantitative descriptions of the behaviour of the following proteins: (* indicates novel biobrick)
*Latherin: a surfactant protein component of horse sweat.
*Ranaspumin-2: a surfactant protein used in the formation of foam nests built by the Tangara frog.
*Phosphodiesterase with VieA domain (PDE): a protein that regulates c-di-GMP levels through cleavage of ester bonds. The VieA domain is specific to c-di-GMP.
Colicin E2 (BBa_K131000): a DNase which triggers cell lysis.
T4 endolysin (BBa_K112806): a lysozyme-like protein produced by T4 bacteriophage in order to lyse the cell and release virions.
Translation
Translation rate calculations were based on average translation rates of E. coli, the following assumptions were made:- RNA transcripts of the protein, once triggered, were generated at an average rate of 40 aa/sec
- each cell contained an average number of ribosomes – 18000 ribosomes/cell
Using these assumptions, the number of ribosomes working on a single RNA transcript was estimated. The rate of protein translation was predicted based on the length of the protein being produced. Also, expression of those molecules is driven by the same ribosome binding site (RBS).
| Name of molecule | Length (a.a.) | Molecular weight (g/mol) | Diffusion rate (mm2/min) | Critical concentration (molecules) |
| Ranaspumin-2 | 98 | 11287 | 1.68*10-4 | 374 |
| T4 endolysin | 164 | 18500 | 1.52*10-4 | 3000 |
| Latherin | 220 | 24400 | 1.38*10-4 | 173 |
| Colicin E2 | 563 | 60000 | 0.72*10-4 | 1629 |
Diffusion
Specific parameters for all of the molecules were initially searched for in the literature (possible diffusion coefficients, size, translation rates and critical concentrations). Due to the lack of specific information and experimentation done on certain molecules, in most cases it was necessary to make additional assumptions and/or use average values to make reasonable estimates of the parameter values operating our system.
The diffusion rate depends on the size of the molecule and the viscosity of the environment. Biofilm is usually very dense and viscous, thus diffusion through it is impaired and slow. We used a one-dimensional diffusion equation to calculate the rate at which modelled molecules should move through the biofilm:

Equation 1
where:
P (r,t) – number of molecules at the certain distance after a certain time
A – total number of molecules in the system
r – distance from point of origin (mm)
D – diffusion coefficient
t – time (min)
Based on the research of Lawrence et al. (1994) and assuming that the diffusion coefficient depends on the molecular weight of the molecule in a linear fashion, we estimated possible biofilm diffusion rates for Latherin, Ranaspumin-2, T4 endolysin and Colicin E2 (Table 1).
For PDE the purpose of the model was to estimate how long it would take to deplete the cell completely of c-di-GMP after expression. It was assumed that each cell contained an average concentration of c-di-GMP and the enzyme was working at its full capacity.
Required calculations and graph plotting were performed with OpenOffice Spreadsheet and GnuPlot software.
The results from models are very informative for guiding our biological engineering of functionalised biofilms. In the case of Colicin E2, research shows [Pugsley, A., 1983] that the protein needs to be expressed for about 90 minutes to cause the cells to lyse. However, molecules like latherin and ranaspumin-2, because of their mode of action, are expected to require at least an hour to cause a desirable effect. For that reason the initial concentrations need to exceed the critical concentrations by a factor 8, so that it will not be diluted by diffusion before being able to affect the area we want to sculpt through the dispersion of biofilm. Since the time of expression of the relevant molecules is controlled by light inducible promoters in our engineered biofilms, concentrations will depend on the timing of the respective light exposure. Estimated times and concentrations are presented in Table 2.
| Name of molecule | Translation rate (molecule/min) | Achieving Critical Concentration (min) | Achieving 8xCritical concentration (min) |
| Colicin E2 | 18.3 | 90 | N/A |
| T4 endolysin | 57.2 | 52 | N/A |
| Latherin | 46.9 | 3 | 29 |
| Ranaspumin-2 | 105.3 | 3 | 28 |
The attainable spatial resolution was calculated based on the diffusion rates of the molecules, i.e. what will be the size of the affected area compared with the area being illuminated. The values of achievable resolution was acquired by calculating at what distance from the initial point we can still observe a concentration, achieved through the diffusion, exceeding the critical concentration for the respective molecule. The initial calculations show that we should be able to achieve an average resolutions as small as ~0.1 mm. The values are presented in Table 3.
| Name of the molecule | The attainable spatial resolution (mm) | The spatial resolution in terms of No. cells |
| Latherin | 0.08 | 40 |
| Ranaspumin-2 | 0.09 | 45 |
| Colicin E2 | N/A | 1 |
| T4 Endolysin | N/A | 1 |
However, while estimating resolution the protein degradation rate should also be taken into consideration. Where we do this, the number of molecules has to be modelled as being time-dependent according to the following equation:

Equation 2
where:
Pf (r,t) – Final number of molecules at given distance at given time
P (r,t) – initial number of molecules according to Equation 1
λ – rate of degradation (molecule/min)
t – time passed (min)
At the moment specific rates of degradation of the molecules in our system are not known, so simulations had to be done for estimated parameters based on general protein degradation rates. In general, our models predict that resolutions will be even better than 0.1 mm, when diffusing molecules are also degraded. Graphical representations of models of diffusion through biofilm for relevant molecules are shown in Figures 1 and 2. All three molecules are shown on a single Figure 3 to allow comparison of their relative diffusion rate through a biofilm.
| (a) Latherin |
 |
| (b) Ranaspumin-2 |
 |
| (c) Colicin E2 |
 |
| (d) T4 Endolysin |
 |
| (a) Latherin |
 |
| (b) Ranaspumin-2 |
 |
| (c) Colicin E2 |
 |
| (d) T4 Endolysin |
 |
 |
For the model of PDE activity we made several simple calculations and assumptions. The concentration of c-di-GMP in an average bacterial cell is low, 1.25x10-17 [μmol/cell], the Vmax of PDE is 1.5457x10-15 [μmol min-1 molecule-1]. This means an average cell can produce around 99.6 molecules of PDE per minute. As a result, fully active PDE should be able to deplete a cell of c-di-GMP in ~0.01 minutes (0.6 secs). Therefore, to compensate for any inaccuracies, allowing production of PDE for a full minute should deprive the cell completely of c-di-GMP. Lack of c-di-GMP would increase motility of bacteria and cause biofilm dispersal. Using this in concordance with surfactant protein expression should generate a cumulative effect on dispersal.
All above models were calculated for a single cell. Because of that and also the fact that for building the models many assumptions and average values were used, the properties of our real engineered system will be slightly different from the predictions shown above. In order to improve the accuracy of our predictions, additional quantitative experiments have to be performed to obtain more precise and specific parameter values. These could then be used to improve the model accuracy for future predictions.
Multi-cell models
Due to the fact that an average biofilm comprises of 3x107 CFU/cm2 and an average E. coli is 2 μm long, the density of cell in any biofilm will be relatively high. Therefore, in order to predict the behaviour of our system when it involves more than one cell, the following models for Latherin and Colicin E2 were created. Both of those models consider a system where there are 21 cells in a row the centres of which are spaced at a distance of 2 μm between each other. Because they all produce the same protein, the overall concentration in the environment is higher than in the single cell situation. That causes the spatial resolution to be poorer, since the concentration exceeds the critical concentration farther from the point of origin than in the single cell models. The difference between the single-cell and the multi-cell models varies, depending on the rate of diffusion and the critical concentrations, but in all cases it is affected. However, a better resolution can be easily acquired by reducing the time the cells are exposed to the specific light wavelength, hence the production of the relevant proteins will be smaller. Therefore, the concentration will not exceed the critical concentration in areas far away from the illuminated parts. That means that the spatial resolution can be precisely controlled by time of light exposure. To establish the optimal time of illumination, a series of experiments need to be conducted. Figures 4 and 5 show the graphical representations of those models.
 |
 |
References:
1. Beeley et al., 1986. Isolation and characterization of latherin, a surface-active protein from horse sweat. Biochemical Journal, 235, pp. 645-650.2. Buchler et al., 2005. Nonlinear protein degradation and the function of genetic circuits. PNAS, 102(27), pp. 9559-9564.
3. Escherichia coli statistics. Institute for Biomolecular Design. Source: internet [http://www.ccdb.ualberta.ca/CCDB/cgi-bin/STAT_NEW.cgi].
4. Goldberg, A. & Dice, J., 1974. Intracellular protein degradation in mammalian and bacterial cells. Annual Review of Biochemistry, 43, 835-869.
5. Kennedy, M., 2011. Latherin and other biocompatible surfactant proteins. Biochemical Society Transactions, 39, pp. 1017-1022.
6. Kaplan, J., 2010. Biofilm dispersal: Mechanisms, Clinical Implications, and Potential Therapeutic Uses. Journal of Dental Research, 89(3), pp.205-218.
7. Konisky, J., 1982. Colicins and other bacteriocins with established modes of action. Annual Review of Microbiology, 36, pp. 125-144.
8. Lawrence et al., 1994. Determination of diffusion coefficients in biofilms by confocal laser microscopy. Applied and Environmental Microbiology, 60(4), pp. 1166-1173.
9. Levi et al., 2011. Cyclic Diguanylate Signalling Proteins Control Intracellular Growth of Legionella pneumophila. mBio, 2(1), e00316-10.
10. Jackson et al., 2001. Growing reproducible biofilms with respect to structure and viable cell counts. Journal of Microbiological Methods, 47, pp. 1-10.
11. Mackenzie et al., 2009. Ranaspumin-2: structure and function of a surfactant protein from the foam nests of a tropical frog. Biophysical Journal, 96, pp. 4984-4992.
12. Mireles et al., 2001. Salmonella enterica Serovar Typhimurium Swarming mutants with altered biofilm-forming abilities: Surfactin inhibits biofilm formation. Journal of Bacteriology, 183(20), pp. 5848-5854.
13. Molecular structures. RCSB Protein Data Bank. Source: internet [http://www.pdb.org/pdb/home/home.do]
14. Pugsley, A., 1983. Obligatory coupling of Colicin release and lysis in Mitomycin-treated Col+ Escherichia coli. Journal of General Microbiology, 129, pp. 1921-1928.
15. Tamayo et al., 2005. The EAL domain Protein VieA is a cyclic diguanylate phosphodiesterase. The Journal of Biological Chemistry, 280(39), pp. 33324-33330.
16. Ursell, T., 2005. Diffusion of solid particles confined in viscous fluid. Source: internet [http://www.rpgroup.caltech.edu/courses/aph162/2006/Protocols/diffusion.pdf].
17. Ursell, T., 2007. The diffusion equation: a multi-dimensional tutorial. Source: internet [http://www.rpgroup.caltech.edu/~natsirt/aph162/diffusion.pdf].
18. Wang et al., 2000. Holins: the protein clocks of bacteriophage infections. Annual Review of Microbiology, 54, pp. 799-825.
19. Webb et al., 2003. Cell death in Pseudomonas aeruginosa Biofilm development. Journal of Bacteriology, 185(15), pp. 4585-4592.
 "
"
