Team:Glasgow/Lovcharacterisation
From 2011.igem.org
LOV2 Purefication From Raw Cell Extract
Aims
Examine properties of the LOV2 protein.Methods
LOV domain purificationPreparation of the calmodulin affinity resin column A calmodulin affinity resin column was set up with a retort stand and partly filled with binding buffer. 2ml of CAM affinity buffer plus a little more binding buffer were added to the column and allowed to flow through to form a packed bed, with the flow through collecting in a glass beaker underneath. Finally the resin was washed with 10ml more of binding buffer, causing as little disturbance to the resin bed as possible. This was to prepare the column for the addition of an E.coli total protein extract containing the PHOT1 LOV2 domain fused to a CBP tag .
Affinity purification of LOV domain Of a 2ml centrifuged sample of protein extract, 1.8ml was loaded into the column (leaving the remaining 0.2ml for SDS-PAGE analysis) and allowed to flow through and collect into a microfuge tube. The liquid in the column turned yellow after this addition as the protein passed through the tube. The column was then subsequently washed with 3ml of binding buffer and any flow through collected in the same tube. The LOV domain was eluted by pipetting 0.5ml of elution buffer into the column and collecting the eluate in a microfuge tube before immediately placing on ice. This was repeated six more times to give a total of seven different labelled eluates, each with different concentrations of LOV domain in them.
Preparation of samples for SDS-PAGE
With the eluates cooling on ice, 12.5µl of the remaining column load, the flow-through fraction and each eluate were mixed with 12.5µl of 2xSDS loading buffer (giving a final volume of 25µl). This was then immediately heated to 100°C to prevent denatured proteins being destroyed by any protease present, then allowed to cool.
Measurement of protein concentration A Bradford assay was then carried out to determine the concentration of LOV protein in the eluates using a sample of each eluate diluted to 1ml with water, 0.9ml Bradford reagent and 0.1mg/ml BSA stock solution, the volume of which was calculated so that the standard curve would cover 0.5-10µg of BSA. A control containing no protein was also included in the standard curve for comparison. Each sample was then mixed and placed in a plastic cuvette and the absorbace of each measured in a spectrophotometer at 595nm. The absorption values of the column load, flow through and eluates were also measured by making a 10-fold dilution of the column load and flow through, then transferring 10µl of these, plus 10µl of each eluate to separate microfuge tubes (in duplicate). 90µl of binding buffer and 900µl of Bradford reagent were also added to each tube before decanting into a plastic cuvette and measuring the absorption in the same way as before. The concentration of LOV protein in each sample was calculated using the standard curve drawn from the BSA absorptions.
Afterwards the samples were illuminated with a UV lamp in a darkroom. Any LOV protein present fluoresced yellow as expected, even though it appeared colourless under normal light conditions. After inspection the samples were placed back on ice to remain cool until an SDS-PAGE gel had been prepared.
Preparation of SDS-polyacrylamide gel
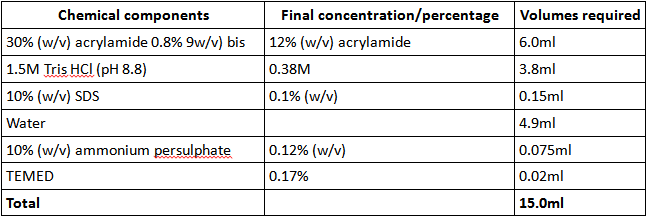
The 12% separating gel was set up by mixing the following ingredients in a conical flask (Table 1):
Table 1: components of 12% separating gel used for SDS-PAGE analysis of LOV protein purity
Chemical components Final concentration/percentage Volumes required
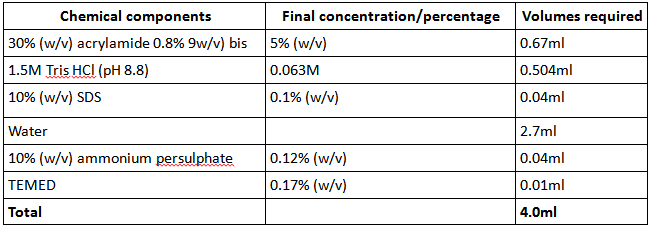
As soon as these components were mixed the gel was poured, then overlayed with water-saturated butanol and left to set. A 5% stacking gel was then prepared in a similar way to the separating gel, this time using the following volumes (Table 2):
Table 2: components of 5% stacking gel used for SDS-PAGE analysis of protein purity
Once the separating gel had set, the water-saturated butanol was removed and the newly prepared stacking gel added in its place. A plastic comb was placed into the gel to create loading wells for the samples to be pipetted into once fully prepared. On solidifying, the comb was removed from the stacking gel and the gel tank was assembled. The tank was then filled with SDS running buffer, and 25µl of each of the purified, defrosted samples was added into each well. Into a final well was added 5µl of pre-stained molecular weight markers (Table 5), then, once connected to the power supply, the gels were run at 80 volts for approximately one hour, until the samples had nearly reached the bottom of the gel.
After the gel had run for the appropriate length of time, the apparatus was dismantled and the gel was placed in a box, ready to be stained.
Staining gels for protein Enough coomassie blue stain was added to cover the gel, then was left for roughly 15 minutes. Water was then used to rinse any excess stain from the gel, keeping the stain for re-use later.
Scanning and analysis
The gel was then carefully placed between 2 sheets of acetate and scanned. The scan was then analysed to see which lanes contained the highest concentration of LOV protein, and which lane had the purest sample. A graph of mobility against log Mr was drawn to estimate the Mr of LOV protein.
 "
"